The Republic of Turkey SELCUK UNIVERSITY
INSTITUTE OF HEALTH SCIENCES
THE DETERMINATION OF SOME PHENOTYPIC AND GENOTYPIC CHARACTERISTICS OF GROUP C STREPTOCOCCUS ISOLATES FROM
DIFFERENT SOURCES
BILAL OSAMAH MOHAMMED MOHAMMED
PhD THESIS
DEPARTMENT OF MICROBIOLOGY (VET)
SUPERVISOR
Prof. Dr. UCKUN SAIT UCAN
ii PREFACE
Today many animals around the world including domestic animals and pets live in close contact with humans and the number has increased in the recent years. The concept of “one health” attracts the attention of researchers and it has become well recognized. In order to keep any new emerging pathogen under control our knowledge should be enhanced to face future challenges such as sudden outbreaks. The zoonotic and opportunistic nature of pathogens such as the Streptococcus genus represents a persistent risk from these bacteria to cause sporadic sever or fatal diseases in various animals at any time.
Streptococcus genus is classified based on its cell wall antigen (A to G). Group C
includes some bacterial species of medical importance that are daily isolated in different laboratories around the world including the microbiology laboratory in Veterinary Faculty of Selcuk University. In Turkey and other parts of the world, information regarding group C Streptococcus is relatively inadequate when it is compared with the information available about other Streptococcus groups.
The aim of our study was the detection of the group C among a number of streptococci isolated from different sources in order to provide more information about this group in Turkey after determining some of its phenotypic and genotypic characteristic.
I would like to thank my professors and education staff and friends in the Microbiology Branch of the Veterinary Faculty for the unlimited support and valuable information they provided. I would like to thank my supervisor Prof. Dr. Uckun Sait UCAN for his support and patience and his good leading. I would like to thank the head of Microbiology Branch Prof. Dr. Osman ERGANIS for giving me this big chance to improve my knowledge in the field of Microbiology and applied sciences. I want also to thank Prof. Dr. Hasan Huseyin HADIMLI who was very supportive during all the years of study. I appreciate the daily instructions and information provided by Associate professor Dr. Zafer SAYIN and Assistant professor Dr. Asli SAKMANOGLU that was very helpful to complete this study. I would like also to thank the Research assistant Mr. Ali USLU for
iii
his motivation and support and helpful information. I thank my amazing family for the trust and unlimited love and support that brought me to this level.
iv CONTENTS
SYMBOLS AND ABBREVIATIONS………..………..vii
Abstract……….………..……..ix
Özet.………..……….x
1. INTRODUCTION ... 1
1.1. General characteristic of Streptococcus genus members... 2
1.2. Group-specific substance (Lancefield classification) ... 3
1.3. Streptococcus Group C ... 4
1.4. The molecular identification of the equine Streptococcus ... 6
1.5. The medical importance of the group C streptococci ... 7
1.5.1. The zoonotic Streptococcus equi subspecies zooepidemicus (SEZ) ... 7
1.5.2. The opportunistic nature of the Streptococcus zooepidemicus ... 8
1.6. The equine health ... 8
1.6.1. The equine IAD ... 9
1.6.2. Equine respiratory infections diagnosis and analysis ... 10
1.6.3. Bronchoalveolar lavage fluid ... 10
1.7. The “one health’’ concept ... 11
1.7.1. Streptococcus group C infections in sheep ... 12
1.7.2. Streptococcus group C and bovine mastitis ... 12
1.7.3. Streptococcus group C infections in poultry ... 12
1.7.4. Streptococcus group C infections in pigs ... 13
1.7.5. Streptococcus group C infections in camels ... 13
1.7.6. Streptococcus group C infections in other domestic animals ... 13
1.7.7. Streptococcus group C human infections ... 14
1.8. Streptococcus group C in Turkey ... 14
1.9. Pathogenicity and Genetic variation ... 16
1.10. Pathogenesis and the genomic era ... 18
1.11. Outbreaks investigation and the importance of the molecular markers ... 19
1.11.1. Random Amplification of Polymorphic DNA ... 20
v
1.12.1. Biofilm detection: ... 22
1.13. Antimicrobials and group C Streptococcus ... 22
2. MATERIALS AND METHOD ... 24
2.1. The subculturing of Streptococci ... 24
2.1.1. Blood agar base formulation (LAB) ... 24
2.1.2. The modified Edwards’s agar formulation (Oxoid) ... 24
2.2. The identification of group C Streptococcus ... 25
2.2.1. Colony characterization ... 25
2.2.2. Gram staining and microscopic observation ... 25
2.2.3. Biochemical identification ... 26
2.2.4. Lancefield grouping latex test ... 27
2.3. The molecular differentiation of the group C Streptococcus... 28
2.3.1. DNA extraction materials preparation ... 28
2.3.2. DNA extraction and purification ... 29
2.3.3. Streptococcus spp. identification ... 29
2.3.4. Streptococcus equi identification and differentiation of its equine subspecies . 31 2.4. Molecular typing and the assessment of the genetic variation ... 32
2.4.1. SZP M-like protein gene detection by PCR ... 32
2.4.2. The sequencing of the SZP gene ... 33
2.4.3. The SZP gene variation assessment ... 33
2.5. Random amplified polymorphic DNA (RAPD) ... 34
2.5.1. The reaction mixture preparation ... 35
2.5.2. The amplification conditions ... 35
2.5.3. The observation of the amplification ... 35
2.5.4. The variation assessment and the dendogram generation ... 35
2.6. Biofilm ... 36
2.6.1. Qualitative Biofilm production assesment ... 36
2.6.2. Quantitative assessment of biofilm ... 38
2.6.3. The biofilm detection by scanning electron microscope ... 41
2.7. The determination of antibiotic sensitivity ... 43
vi
3.1. Colony morphology and Gram staining results ... 44
3.2. The biochemical tests result ... 46
3.2.1. Latex grouping test result ... 46
3.2.2. Sugar fermentation result ... 46
3.3. Molecular identification ... 49
3.3.1. Genus specific PCR results ... 49
3.3.2. The results of the species specific PCR using SodA primer ... 50
3.3.3. Molecular detection of the specific SEM gene of Streptococcus equi subsp. equi ... 51
3.4. Molecular typing and assessment of variation result ... 52
3.4.1. The results of SZP gene detection by PCR ... 52
3.4.2. SZP gene sequencing results ... 54
3.4.3. The phylogenic tree based on the variation of SZP gene sequence ... 60
3.5. Random amplified polymorphic DNA (RAPD) result ... 60
3.5.1. Phylogenic tree and grouping based on RAPD results ... 62
3.6. Biofilm formation ... 63
3.6.1. Congo red agar method results ... 63
3.6.2. Evaluation of the biofilm degree by crystal violet staining method ... 65
3.6.3. Biofilm detection by SEM ... 69
3.7. Antimicrobial sensitivity testing results ... 71
4. DISCUSSION ... 75
5. RESULTS AND SUGGESTIONS ... 81
vii SYMBOLS AND ABBREVIATIONS
SEM Streptococcus equi M-like protein BHI Brain heart infusion
TSB Tryptic Soy Broth OD Optical density
ATCC American Type Culture Collection SODA Superoxide dismutase
mm Millimeter µl Microliter cm Centimeter P Penicillin T Oxytetracyclin
AMC Amoxicillin - Clavulanic Acid KF Cephalothin
SAM Ampicillin-Sulbactam
SXT Sulfamethoxazole-Trimethoprim
SZP Streptococcus zooepidemicus M-like protein SEZ Streptococcus equi subsp. zooepidemicus RAPD Random amplified polymorphic DNA PCR Polymerase chain reaction
viii TRIS Trisaminomethane
SDS Sodium dodecyl sulfate TAE Tris-Acetate-EDTA buffer
CLS Clinical and Laboratory Standards Institute EDTA Ethylenediaminetetraacetic acid
BAL Bronchoalveolar lavage CNE Collagen adhesion in Streptococcus CNA Collagen adhesion in Staphylococcus AST Antimicrobial susceptibility test PBS Phosphate buffer solution
ix ABSTRACT
The Republic of Turkey SELCUK UNIVERSITY
INSTITUTE OF HEALTH SCIENCES
The Determination Of Some Phenotypic And Genotypic Characteristics Of Group C
Streptococcus Isolates From Different Sources
Bilal Osamah Mohammed MOHAMMED Microbiology Department (VET)
PHD THESIS/KONYA-2019
The aim of this study was the detection of group C streptococci in 115 isolates from the laboratory collection of Microbiology Department of the Veterinary Faculty, Selcuk University that had been collected from different sources and the determination of some of their phenotypic and genotypic characteristics. This included the evaluation of biofilm production, susceptibility to different antibiotics, detection of SZP virulence factor gene and the determination of sequence variation among strains and genotyping by RAPD method. Streptococcus equi subspecies zooepidemicus ATCC 35246 was used as a standard strain.
In this study, 41 of the isolates were confirmed to be group C streptococci by using Latex Streptococcus grouping kit. The identification was completed by both biochemical and molecular methods. The whole strains were Streptococcus equi subspecies zooepidemicus. The biofilm formation was evaluated by three different methods (congo red agar, crystal violet staining method and SEM microscope). In spite of the slight incompatibility among the congo red agar and crystal violet staining methods the majority of isolates were confirmed as strongly biofilm producing strains. The testing of antimicrobial susceptibility revealed a relatively high sensitivity to five antibiotics and mild resistance to the sixth antibiotic. The virulent gene SZP was detected in 35 of the isolates including the reference strain ATCC 35246. The SZP sequence was confirmed to be variable among different strains after comparing all the sequences and generating the phylogenic tree. The genotyping by RAPD method revealed the high variability among strains and also similarity between some of them. In conclusion, group C streptococci originating from the horse respiratory system were found variable.
Keywords: Streptococcus equi subspecies zooepidemicus, Latex grouping, biofilm, antimicrobial
x ÖZET
T.C.
SELÇUK ÜNİVERSİTESİ Sağlık Bilimleri Enstitüsü
Farklı kaynaklardan Izole Edilen C Grubu Streptokokların Bazı Fenotipik Ve Genotipik Özelliklerinin Belirlenmesi
Bilal Osamah Mohammed MOHAMMED Mikrobiyoloji (VET) Anabilim Dalı
DOKTORA TEZİ / KONYA-2019
Bu çalışmanın amacı, farklı kaynaklardan toplanan Veteriner Fakültesi, Mikrobiyoloji Anabilim Dalı laboratuvar koleksiyonunda bulunan 115 izolatta C Streptococcus grubunun tespit edilmesi ve bazı fenotipik ve genotipik özelliklerinin belirlenmesidir. Bunlar, biyofilm üretiminin değerlendirilmesi, farklı antibiyotiklere duyarlılık, SZP virülans faktörü geninin tespiti ve suşlar arasında sekans varyasyonunun belirlenmesi ve RAPD metodu ile genotiplemedir. Standard suş olarak Streptococcus equi subspecies zooepidemicus ATCC 35246 suşu kullanıldı.
Bu çalışmada, lateks Streptokok gruplama kiti kullanılarak izolatların 41'inin C grubu Streptokok olduğu doğrulandı. Tanımlama hem biyokimyasal hem de moleküler yöntemlerle yapıldı. Suşların tamamı Streptococcus equi subspecies zooepidemicus'du. Biyofilm oluşumu üç farklı yöntemle değerlendirildi (kongo kırmızı agar, kristal violet boyama yöntemi ve SEM mikroskobu). Kongo kırmızısı agar ve kristal violet boyama yöntemleri arasındaki düşük uyuşmazlığa rağmen, izolatların çoğunun kuvvetli biyofilm üreten suşlar olduğu doğrulandı. Antimikrobiyal duyarlılık testi, denenen antibiyotiklere nispeten yüksek duyarlılık ve bir ı antibiyotiğe hafif direnç gösterdi. Virülent gen SZP, ATCC 35246 referans suşu dahil izolatların 35'inde tespit edildi. SZP dizisinin, bütün dizileri karşılaştırdıktan ve filogenik ağacı oluşturduktan sonra farklı suşlar arasında değişkenlik olduğu doğrulandı. RAPD yöntemiyle genotiplendirme, suşlar arasındaki yüksek değişkenliği ve ayrıca bazıları arasındaki benzerliği ortaya koydu. At solunum sistemi kökenli C grubu streptokoklarda varyasyon olduğu kanaatine varıldı.
Anahtar Sözcükler: Streptococcus equi subspecies zooepidemicus, Lateks gruplaması, biyofilm,
1 1. INTRODUCTION
Streptococcus genus members are daily isolated in laboratories from various areas
around the world as the causative agent of various local and generalized infections in humans and other animals. Strangles and mastitis are considered the main and most important animal diseases caused by this bacterial group. Streptococcus is also the main cause of a wide range of infections in humans, including erysipelas, cellulitis, and pharyngitis.
The Streptococcus genus is classified as serogroups (A to G). This classification was established based on the surface antigens carried by each group members. Some of the Streptococcus members do not carry one of these designated surface antigens and classified as not groupable. Streptococci belong to the group C include two closely related species the Streptococcus equi subspecies zooepidemicus which is a worldwide distributed and commensal of the upper respiratory tract of healthy horses and the
Streptococcus equi subspecies equi the causative agent of the highly contagious fatal
equine disease the strangles. The third member of group C is Streptococcus dysgalactiae subsp. equisimilis is considered of less importance. Its normal habitat in the horse appears to be the skin and mucous membrane. Streptococcus zooepidemicus and Streptococcus
equisimilis can cause opportunistic infections in various animal species including
humans.
The infectious respiratory tract diseases in horses appear in acute and chronic forms. These infections affect the function of the respiratory system causing a significant decrease in the horse’s performance which represents a major economic problem. There is a lack of sufficient amounts of information about the incidence of bacterial species and subspecies of the horse’s respiratory tract when it is compared with other species.
The zoonotic nature of the Streptococcus has been a major source of risk of infections. The direct contact between animal breeders and their domestic animals make them exposed to the transmition of a variety of pathogens. The contamination of animal products has been one of the major causes of dieases outbreaks around the world.
2 1.1. General characteristic of Streptococcus genus members
The genus Streptococcus is worldwide in distribution. This bacterial genus includes relatively harmless members of the oropharyngeal flora in animals. Most of the streptococci inhabit the mucosa of the upper respiratory and urogenital tracts of human and other animals of veterinary interest. They are usually susceptible to desiccation and cannot survive for long outside the animal host (Ryan and Ray 2004).
Staphylococcus is the most frequently isolated bacteria from mastitis in sheep.
The second group of bacterial agents responsible for sheep mastitis is the Streptococcus (Uçan 2002). This includes the most frequently identified species Streptococcus
agalactiae, Streptococcus uberis, and Streptococcus dysgalactiae (Uçan et al. 2005).
The gram positive streptococci are facultative anaerobic with complex nutritional requirements and no catalase activity. They are 2μm spherical in shape and divides in one plane. This results the known strip like liner array form of the Streptococcus genus. They are adapted as commensals or parasites in various species of the animal kingdom.
Streptococcus is very well known and familiar to all laboratories around the world. The
identification usually starts by observing the colony characteristics and hemolytic properties. The differentiation of the Streptococcus is usually based on the carbohydrate and protein antigen composition and also fermentation. The DNA sequencing of 16 and 23S rRNA genes is usually useful for molecular identification for most of the streptococci (Gyles et al. 2010).
Table 1.1.The classification of the Streptococcus.
Group Phylum Class Order Family Genus
3 Figure 1.1. Streptococcus under microscope. (Ryan and Ray 2004).
At the beginning of the 20th century the classification of the Streptococcus was based on the biochemical tests that were sufficient to associate a small number of the streptococcal pathogens of animals and human. The laboratory differentiation had depended on these tests until Rebecca Lancefield demonstrated carbohydrate antigens (A to G) that became later the main tool for the classification and taxonomy of the hemolytic and also non-hemolytic streptococcal species. The demonstration of these carbohydrates which are actually virulent segments of the streptococcal genus brought in a new taxonomic group called the pyogenic streptococci (Ryan and Ray 2004).
1.2. Group-specific substance (Lancefield classification)
The cell wall of Streptococcus contains the carbohydrates that are the basis of serologic grouping into Lancefield groups. The serologic specificity of the group specific carbohydrate is determined by an amino sugar molecule. In group A it is a rhamnose-N-acetylglucosamine while for group B it is rhamnose-glucosamine polysaccharide and for group C it is rhamnose-N acetylgalactosamine. In group D it is glycerol teichoic acid that contains D-alanine and glucose molecules. And group F contains glucopyranosyl-N-acetylgalactosamine (Jawetz 2007).
4 Figure 1.2. The antigen structure of Streptococcus cell. (a) Capsule is hyaluronic acid
(b) Cell wall protein antigens M, T, and R. (c) Group-specific carbohydrate is rhamnose N-acetylglucosamine (Jawetz 1989).
1.3. Streptococcus Group C
The group C members of genus Streptococcus can be easily differentiated. The first step is noticing the very clear beta hemolysis surrounding the gram positive streptococci colonies on blood agar. Latex agglutination is commercially available and commonly used grouping test. Specific C-substances antisera are used to determine the group to which the streptococcal isolates belong. Latex particles in suspension are coated with specific antibodies represent each group A to G of Streptococcus. The latex test includes reagents for enzymatic and acid extraction of the antigen. One drop of the extracted antigen suspension is mixed with one drop of the latex-antibody suspension. The agglutination usually occurs within one minute after mixing (Quinn et al. 2011).
5 Figure1.3. Diagrammatic representation of latex agglutination test used for Streptococcus identification (Quinn et al. 2011).
Currently group C Streptococcus include three main species Streptococcus equi subspecies equi, Streptococcus equi subspecies zooepidemicus and Streptococcus
dysgalactiae subsp. equisimilis that had been added recently to the group. This was due to
the presence of the same group C carbohydrate antigen. The genetic similarity between the former human isolates to the animal group C strains of Streptococcus dysgalactiae subsp. equisimilis led to their placement in the same species but to different subspecies (Koneman 2011).
The mucosal commensal of the equine oral cavity, pharynx and respiratory tract
Streptococcus equi subspecies zooepidemicus is the most frequently beta-hemolytic
isolated streptococci from horses. Streptococcus dysgalactiae subsp. equisimilis is the less frequently isolated member of the group C. The third member of this group is the host restricted highly infectious pathogen the Streptococcus equi subspecies equi. Studies have shown that many horses are infected in some stage and carry these bacteria in their guttural pouch but only small percentage of susceptible horses are in a risk of developing disease (Erol et al. 2012).
A short range of sugar tests includes lactose, sorbitol, trehalose and maltose is usually used for the further differentiation among the three members of group C equine streptococci Streptococcus equi subspecies zooepidemicus, Streptococcus equi subspecies
6 Table1.2. Differentiation of equine group C streptococci by sugar fermentation.
v = variable reactions, (-) = a few strains are negative (Quinn et al. 2011).
Species Lactose Sorbitol Trehalose Maltose
Streptococcus zooepidemicus + + - +(-)
Streptococcus equi - - - +
Streptococcus equisimilis v - + +
1.4. The molecular identification of the equine Streptococcus
The identification of the equine Streptococcus traditionally relied on the phenotypic characteristics which are the C-specific antisera and sugar fermentation. The development in nucleic acid technology and the analysis of the 16rRNA resulted new methods that can be used for more specific bacterial identification of several species. But in case of the Streptococcus equi the 16rRNA was found useless for the species specific identification. This was after the sequencing of the internal parts of the 16rRNA gene of both Streptococcus equi subspecies zooepidemicus and Streptococcus equi subspecies
equi. The two closely related species share an identical sequence which will not allow a
specific differentiation. The sequence variation among Streptococcus equi subspecies
zooepidemicus strains is another reason for considering it useless to choose this sequence
and depend on it as a target for the differentiation of the two species. Several specific differentiation systems have been designed in order to find a rapid and efficient way for the differentiation of the group C Streptococcus members. Depending on previous studies a PCR protocol had been developed and used efficiently. The SEM gene encodes for the M-like protein exclusively found in Streptococcus equi subspecies equi was used for its identification while the superoxide dismutase (sodA) gene was used for the species specific identification of the Streptococcus equi subspecies zooepidemicus. Another more efficient PCR multiplex system for rapid identification was suggested. In addition to the sodA gene two more genes were selected for further identification. These genes encode for exotoxins seeH, seeI found only in Streptococcus equi subspecies equi (Alber et al. 2004; Javed et al. 2016).
7 1.5. The medical importance of the group C streptococci
The β-hemolytic Streptococcus equi subspecies zooepidemicus can infect various domestic animals other than horses such as cattle, sheep, goats, pigs, dogs, and cats. The other closely related species, the host specific Streptococcus equi subsp. equi causes strangles which is a well-known and highly contagious and serious disease in horses. The two members of the Lancefield group C Streptococcus equi and Streptococcus
zooepidemicus share around 98% DNA sequence homology. S. zooepidemicus share also
>80% DNA sequence homology with the human streptococcal pathogen, Lancefield group A member the Streptococcus pyogenes. The streptococci group C and A share also some of the main virulence factors essential to initiate infections such as the anti-phagocytosis M-like proteins, super antigens (sAgs) and also the hyaluronic acid found in the capsule of certain strains. Although the human infection with S. zooepidemicus is considered rare, and most of the published reports are back to the 1980s, there are still occasionally reported human infections as a result of unpasteurized milk or homemade cheese consumption. In animals Streptococcus dysgalactiae subsp. equisimilis infections can also occur. There have been some reports about bacteremia, endocarditis, pneumonia and also toxic shock syndrome caused by this beta hemolytic Streptococcus. It has attracted the attention recently after being isolated from abscessed lymph nodes in some strangle like cases. It has also shown its potential to cause abortion in some domestic animals (Preziuso et al. 2010).
1.5.1. The zoonotic Streptococcus equi subspecies zooepidemicus (SEZ)
The Lancefield group C member the Streptococcus equi subspecies
zooepidemicus is a commensal organism of the tonsil and nasopharyngeal mucosa of the
equine. This species may opportunistically start respiratory infections such as purulent rhinitis, bronchitis and pneumonia in foals and donkeys of different ages. In older horses that have been under heat or transportation stress, it may also cause acute hemorrhagic pneumonia (Sellon 2013).
A study was conducted in order to determine and identify the various β-hemolytic streptococci species existed in 2,391 isolates from lymph nodes, placenta, fetal tissues,
8
and genital tract and provide a comprehensive picture as an example of their prevalence and distribution. The isolated species percentage was Streptococcus equi subsp.
zooepidemicus (72.0%), Streptococcus dysgalactia subsp. equisimilis (21.3%), Streptococcus equi subsp. equi (5.8%) and streptococci spp. (0.9%) (Erol et al. 2012).
1.5.2. The opportunistic nature of the Streptococcus zooepidemicus
A study for comparing the phenotypes of Streptococcus zooepidemicus isolated from tonsils of healthy horses with other isolates obtained from foals and donkeys with pneumonia was conducted. The results showed that tonsils of each healthy horse were colonized by several phenotypes similar to those of foals or donkeys with pneumonia. This shows the opportunistic nature of the S. zooepidemicus for causing serious lung and airways infections (Anzai et al. 2000).
1.6. The equine health
The Horse races and horse breeding sector is economically important. Small airway inflammatory diseases have been a main problem that causes poor performance and exercise of horses in various breeding areas around the world. Along with cough and exercise intolerance, the bacterial infections would be confused with viral and also fungal infections. Further diagnosis is usually necessary to confirm whether the dysfunction is caused by an infection. Bronchoalveolar Lavage fluid (BAL) has been considered as an efficient method for specimen collection from horses especially when chest radiography is not available in the field. The recovery of microbiologic samples allows more understanding of the causative agents that would contribute to any condition (Hoffman 1997).
Horse riding sports activities are growing in many countries around the world. The high risk of transmission and the severe illness caused by S. zooepidemicus infection emphasize the idea of considering it as a serious emerging zoonotic pathogen. The direct contact with horses increases the risk of transmission of the equine opportunistic commensal to cause a severe infection in humans. A published article showed clearly that risk when isolates from different 3 cases of patients with severe infections in eastern Finland were analyzed. As horse trainers and breeders, these 3 patients were in direct
9
contact with horses that had not shown any clinical signs of respiratory illness or any other diseases during examination. The isolates were analyzed by using pulsed-field gel electrophoresis, multilocus sequence typing, and sequencing of some virulence gene. The molecular typing methods used in this study showed that human and equine isolates were identical or closely related (Pelkonen et al. 2013).
A retrospective clinical study described a number of ulcerative keratitis cases in horses that had been caused by the Beta hemolytic Streptococcus subsp. zooepidemicus. These bacteria were isolated from horses with aggressive cases of ulcerative keratitis. The treatment required therapeutic agents and surgical intervention in the majority of the cases. The treatment was challenging and some of the infected eyes did not retain its function completely (Brooks et al. 2000).
1.6.1. The equine IAD
The inflammatory airways disease (IAD) is the inflammation that is detected in the trachea and bronchi. The (IAD) has the greatest impact on the health of horse’s populations particularly in the young thoroughbred race horses. The mechanism of IAD development in horses is poorly defined. It is thought that a variety of etiological agents such as housing, feeding, climate, the use of preventive medicine practices contribute and make different horse populations susceptible to the development of the disease (Couëtil et al. 2007; Wood et al. 2005a).
In races respiratory disease is considered as the second most common causative of lost training days and training disruption before competitions and this was the reason for performing many studies about young race horses. Studies have shown variation among different yards and also age groups. Researchers concluded that respiratory diseases are more common in young racehorses and it decreases significantly with age. Some studies also showed that the bacterial isolates as primary infectious agents were more common than viral agents in causing respiratory diseases (Wood et al. 2005b).
The normal flora of the equine respiratory tract and oral cavity play a major role in the health of various organs and tissues such as skin, gastrointestinal tract, urogenital
10
system and also the respiratory system. The upper respiratory tract of horses is usually inhabited by a variety of aerobic and anaerobic species that compete with invading pathogens. In healthy horses it prevents their colonization to the epithelial tissue when they present in large numbers. This includes Streptococcus spp., Streptococcus equi subsp. zooepidemicus, Pasteurella spp., Escherichia coli and Actinomyces spp. and
Mycoplasma felis. Culturing of tracheobronchial aspirate from horses that have been
under stress usually results the growth of various bacteria and fungi as well. The source of these species along with the mucus accumulation is usually the result of contamination from the upper respiratory tract (Sellon 2013; Mete 2015).
Studies have shown the link between the respiratory inflammation and bacterial infections in several cases. They may appear as primary secondary infections after viral diseases causing the contagious infections that may result in chronic respiratory diseases, impaired pulmonary function and also exercise induced hemorrhage in active race horses. Other studies have actually shown no association between the bacterial isolates and the outcome of the disease (Carman et al. 1997; Racklyeft and Love 2000).
1.6.2. Equine respiratory infections diagnosis and analysis
The respiratory fluid analysis and microbiological diagnostic techniques have improved considerably which allows better interpretation of results in order to understand the pathophysiological mechanisms of various pathogens. Care should be always taken during the interpretation of bacterial cultures obtained from airways aspirates. Quick conclusions about any bacterial isolates being the pathogenic agent during respiratory disease should not be accepted as infections may be confused with the colonizing normal flora or the transient airway contamination (Richard et al. 2010).
1.6.3. Bronchoalveolar lavage fluid
There are a number of ancillary diagnostic tests available for the accurate detection and identification of subclinical horse diseases. Bronchoscopy is one of these techniques which are mainly useful in the observation of trachea and bronchi. Other methods are employed for the identification of infectious agents such as nasal swabbing, tracheal aspirate, pleural effusion and bronchoalveolar lavage fluid. In cases of profuse
11
nasal discharge and cough the rapid method of nasal swabbing is usually used. The culturing of the nasal discharge would result microbes from the lower respiratory tract in case of pulmonary diseases but it usually contains contaminants from the upper respiratory tract. Although tracheal aspirate and pleural effusion can provide useful information it cannot represent the small airways cytology. Equine pulmonary diseases involve the smaller bronchioles before it diffuse throughout the lung. BAL is increasingly used as it allows the sample collection from the various distal segments of the lung and other parts of the respiratory tract (Dixon 1997).
Bronchoalveolar lavage (BAL) method allows deciding on the suitable treatment and also allows researchers to determine the nature of the recovered inflammatory cells by BAL such as neutrophils, eosinophils and mast cells. Unlike humans, horses usually tolerate this procedure of sample collection. The procedure includes passing a sterile lubricated tube through the ventral meatus of a sedated horse into the pharynx until it reach a particular depth inside the respiratory tract. The next step includes flushing and aspiration of a warmed sterile saline solution containing some kind local anesthetic with slight effect in order to bring back cells into the fluid. The fluid is then collected in a sterile plastic bottle or syringe (Hoffman and Viel 1997).
1.7. The “one health’’ concept
The infections transmission can occur directly through the physical contact or sufficient proximity between two animals. The indirect transmission of infectious agents to animals is usually by the exposure to contamination. Materials carrying pathogens transmitted from an infected host have been the sources of all outbreaks in various investigation studies. This includes contact with animal exudates such as feces, urine and nasal discharge. The identification of transmission routes is extremely difficult without the molecular typing methods of biotechnology (Pisoni et al. 2009).
12 1.7.1. Streptococcus group C infections in sheep
It has been also reported that another less frequently appearing species such as
Streptococcus parasanguinis and Streptococcus zooepidemicus were also isolated from
mammary glands of sheep. A study described an unusual outbreak of clinical mastitis in sheep that emphasized the possibility of sporadic occurrence of infections caused by the normal flora of the respiratory and urogenital tracts of equines the Streptococcus
zooepidemicus in sheep. A sheep flock and after contamination from a single donkey with
no signs of any clinical infections and in spite of conditions and the hygiene which were considered up to the standards, an infection detected and was first confused with
Streptococcus dysgalactiae infection. Due to the similar clinical signs it was called
pseudoagalactia. The infection was actually by the equine Streptococcus zooepidemicus. The study showed the relatively high morbidity rate of 22% that affected the milk production seriously and also showed the high resistance to tetracycline after treatment failure (Las Heras et al. 2002).
1.7.2. Streptococcus group C and bovine mastitis
A study was conducted in order to investigate the bacterial agents that caused mastitis in a number of dairy cows from different states in Nigeria. Different bacterial species were isolated from 307 milk samples. Streptococcus zooepidemicus made up 3.85% of the isolates (Adesola 2012).
1.7.3. Streptococcus group C infections in poultry
Poultry industry can be considered as one of the most important sectors in animal production. Sudden disease outbreaks have always affected the chicken breeders economically. Chicken infection with Streptococcus zooepidemicus is rare but there have been some reports of serious infections of this bacterium that caused the expiry of big numbers of chickens. An outbreak affected 11,000 chickens at the age of 47 weeks was reported. The infection started acute and then developed to chronic stages causing 80% mortality within few weeks. The molecular typing methods used in the epidemiological investigation of the outbreak were very essential in detecting the source on the pathogen.
13
The study showed that the origin of this pathogen was actually a horse that used to be kept in the farm in the same area (Bisgaard et al. 2012).
1.7.4. Streptococcus group C infections in pigs
In China, Streptococcus zooepidemicus is considered the second most important pathogen of swine diseases caused by Streptococcus. A pandemic in the city of Sichuan caused by Streptococcus zooepidemicus led to the death of around 300,000 pigs in 1975. That caused a big economic lose and until now the pig industry is still suffering from this disease (Fan Hong-jie 2009).
1.7.5. Streptococcus group C infections in camels
Camels are economically important in some parts of the world. The camel’s milk is consumed in many countries. Streptococcus equi subsp. zooepidemicus may also cause mastitis and other infections in camels. A published article explained the isolation of a number of beta-haemolytic Streptococci from camel’s milk samples that were collected from different herds and districts in Kenya and Somalia and were identified as
Streptococcus equi subsp. zooepidemicus (Younan et al. 2005).
1.7.6. Streptococcus group C infections in other domestic animals
The increasing number of different kinds of pets around the world attracts the attention to the high risk after several reports about infections that were linked to pets. The equine commensal organism Streptococcus zooepidemicus has been associated with various respiratory and suppurative infections in many pet animals such as dogs, pigs, gerbils, monkeys, minks and also guinea pigs. For instance a family cluster case of purulent infections in Virginia USA was identified and linked to the direct contact with guinea pigs. Until now, a single case of transmission from dog to a handler has been described. This case was analyzed using molecular methods and the results showed that the isolates from the handler and the dog were similar. Another studies about the possible risk of zoonotic infections showed that dogs are considered to pose a higher risk of infection with a Streptococcus zooepidemicus strains more virulent than those carried by horses. Dog were also seen to shed relatively large quantities of these bacteria and also
14
produce more amounts of nasal discharges that keep the humans exposed to more skin and respiratory infections (Abbott et al. 2010; Gruszynski et al. 2015).
1.7.7. Streptococcus group C human infections
Although rare in humans, Streptococcus zooepidemicus infections have been reported as the main cause of complications and death in elderly people and it may be confused with some other conditions such as pyrexia and Listeriosis. The main rout of infection was the ingestion of infected dairy products representing third of the cases. Ten cases of infections through inhalation have been reported. Inoculation as a result of accidents was also a postulated rout of infection in four other cases. The most common complication was the persistent hearing loss and in spite of the treatment with aminoglycosides, it occurred in 19% of the cases. Endocarditis, endophthalmitis, tetraparesis and aphasia, seizures have been also reported and the recovery rate of 38% of cases was significantly low. Some authors show the significant risk of death as a result of complications or the development of secondary infection such as Clostridium. The crude mortality in patients who all died over 70 years old associated with meningitis (which had been considered rare) was 24%. Another authors reported highest death rates of 25-40% with group C streptococcal bacteremia (Eyre et al. 2010).
1.8. Streptococcus group C in Turkey
Although its infections are very limited in Turkey, some cases have been reported and some studies have been conducted regarding Group C Streptococcus. Two cases of infection with group C members’ streptococci were reported and published. A case of fatal meningitis caused by Streptococcus zooepidemicus (Ural 2003) and a case of prosthetic joint septic arthritis caused by Streptococcus equisimilis (Sipahi 2008). Another study for the evaluation of antibiotic susceptibility of 68 group C and 37 group G streptococci that had been isolated between 1995 and 2002 was conducted (Ergin 2003).
A case of abortion due to Streptococcus zooepidemicus infection in mare was reported in Turkey. The histopathological examination of the aborted equine fetus
15
showed mild cellular infiltration of neutrophils, lymphocytes and plasma cells in the lamina propria of the intestine. The microbiological examination showed bacterial growth in the intestine and the liver. Pure Streptococcus zooepidemicus was isolated from the infected organs (shown in figure 1.4.) as the main causative of death (Kocabiyik 2005).
Figure 1.4. (1) Bacterial colonies (arrows) and mild cellular infiltrates (arrow head) in the lamina propria of the intestine. (2) Bacterial emboli (arrows) without inflammatory reaction in the sinusoids of the liver (Kocabiyik 2005).
A study was conducted in Turkey described a case of systemic infection in 22 days old female foal with Streptococcus zooepidemicus. The foal had been suffering from severe pneumonia and also painful swellings in the carpal and tarsal joints. After histopathological examination the microbiological examination of the samples from the foal and the dam revealed clinical mastitis that was caused by Streptococcus
zooepidemicus. The infected milk was seen thick and yellow. It was concluded from the
clinical and microbiological examination that the mastitis led to the infection and death of the foal (Kocabiyik 2008). Another study showed that S. zooepidemicus played an important role in the etiopathogenesis of lower respiratory tract disorders in race horses (Kasap et al. 2018).
16 Figure 1.5. Section of liver during postmortem histopathological examination containing
numerous disseminated, raised, round to oval abscesses of various size caused by
Streptococcus zooepidemicus replacing the liver parenchyma (Kocabiyik 2008).
Another study was conducted to investigate the prevalence of different groups of
Streptococcus in pediatric acute tonsil pharyngitis in Turkey. 200 samples were collected
from children between 1-15 years old. Group C Streptococcus made up 3% of the streptococcal isolates that caused the tonsil pharyngitis (Tasar 2015).
Another study was published. It described a severe case included meningitis, cerebral and paravertebral abscess and arthritis in addition to prosthetic valve endocarditis of a 65 years old patient due to an infection with equine Streptococcus. The patient had been exposed to the risk of infection due to the consumption of dairy products and direct contact with domestic animals and their products. The case required surgical and medical intervention (Kutlu 2018).
1.9. Pathogenicity and Genetic variation
The ability of the equid’s pathogens Streptococcus equi and Streptococcus
zooepidemicus to establish an infection has been explained in a number of studies. These
two species share the majority of their proteins and virulence factors that determine their relationship with their host animals. There is a minor difference between the genomes of these putative ancestral biovars resulted some major differences in the mechanism of pathogenicity. There is still lack of adequate information about the third member of the Lancefield C Streptococcus dysgalactiae subsp. equisimilis which share a low DNA
17
homology with the other two members. Lancefield group C, G and A streptococci express similar groups of fibrous proteins on their surface that can bind to the host matrix and serum proteins. It forms an α-helical coli allow the bacterial cell to bind to immunoglobulin, fibrinogen, kininogen and to other molecules such as the albumin and plasminogen. Studies have shown that this ubiquitous protein that is named M-like protein could be involved in the phagocytosis resistance and evasion of host defenses. It is also thought that they play an important role in adhesion and invasion (Meehan et al. 2001).
The expression of five different wall associated proteins has been detected in different strains of the group C Streptococcus member Streptococcus equi. These proteins are two fibronectin, α2-macroglobulin, immunoglobulin, albumin binding proteins (Lindmark and Guss 1999). Researchers have also mentioned that the most dominantly expressed wall associated finbrinogen binding proteins in the virulent Streptococcus equi are those termed as SEM and SzPse. SzPse is an antigenically cross reactive to the hyper variable surface protein Szp produced by the closely related species Streptococcus
zooepidemicus. Although they share the majority of their genome sequence and proteins,
immunization against Streptococcus zooepidemicus does not protect against
Streptococcus equi (Timoney et al. 1997; Meehan 1998).
Studies have shown that the ongoing bacterial evolution and especially pathogens plays a major role in the development of diseases in animals including human. An important aspect which has not been importantly considered is the genetic material exchange among bacteria that would have shaped the evolution of the host restricted pathogens. Streptococcus equi is a horse restricted pathogen that shares the majority of its genome sequence with Streptococcus zooepidemicus and Streptococcus pyogenes. This was explained in a study conducted by a group of scientists from different universities around the United Kingdom had come with significant results about the genetic transformations led to the evolution of Streptococcus equi as a separate species from its closely related Streptococcus zooepidemicus. The analysis after sequencing and comparison of genomes of isolates from around the world uncovered the genetic events led to the host restricted pathogen emergence. The transformations were the functional
18
loss due mutations, deletions, bacteriophage acquisition of a phospholipase A2 toxin and four super antigens and also a novel iron acquisition system similar to the high pathogenicity island of Yersinia pestis (Holden et al. 2009).
1.10. Pathogenesis and the genomic era
In epidemiological studies it is necessary to identify the bacterial strains correctly in order to improve our understanding of pathogenicity. The conventional biochemical methods can be used for the identification of Streptococcus species even though it is considered a time consuming, doubtful and also depend on the availability of perfectly prepared bacterial cultures. There is a variation in the results obtained from different laboratories. This makes the results need further clarification especially the phenotypic identification to the species level. The improvement of the molecular methods and automation make it a perfect alternative as it is fast, no need for pure culture and makes it possible to identify even dead bacterial strains (Moriconi et al. 2017).
The development of the genomic sequencing and the availability of enormous amounts of information have revolutionized the science of pathogenesis. Understanding of the basic aspects of genes contents that contributes to virulence and host response allows us to generate global patterns to explain the complex regulatory networks that determine the development of diseases (Medini et al. 2008).
There is a rapid progress for the identification and characterization of the unique proteins expressed by the Streptococcus equi the causative agent of strangles in order to accelerate the determination of new vaccine targets. Some of these proteins are expressed on the surface such as the antiphagocytic SEM protein while the others are secreted such as the pyrogenic auperantigen SePE-1 and H. Although it has been frequently isolated from various animals and from horses as the causative agent of respiratory infections and metritis, the available research studies about Streptococcus zooepidemicus are relatively less. The given name “zooepideicus’’ actually implies that this species as a pathogen is not adapted to any host. Non antigenic hyaluronic acid capsule, hyaloronidase, streptolysin S, streptokinase, IgG Fc-receptor, capsule, peptidoglycan and leukocidal toxin are the identical virulence factors of the two closely related equine streptococci.
19
Detecting the virulence factors and their regulatory proteins at the molecular level in different isolates would allow more understanding of the opportunistic behavior of the pathogens (Timoney 2004).
A group of researchers detected a potential virulence factor called CNE. The gene sequence of this collagen-binding protein was identified using bioinformatics tools. Researchers constructed a recombinant molecule containing this gene in order to evaluate its function. This gen was detected in both species Streptococcus equi subsp. equi and
Streptococcus equi subsp. zooepidemicus as well. The similarity between this protein and
the well-known Staphylococcus CNA virulence factor induced the researchers to study its complete function as a novel virulence factor (Lannergård et al. 2003).
1.11. Outbreaks investigation and the importance of the molecular markers
Although Streptococcus zooepidemicus have been reported as the causative agent of meningitis and mastitis in goats, it is not known to be a common isolate from genital infections. Most of laboratories usually do not complete the identification of the isolated bacterial strains from clinical samples. Prior identification or typing only to the group level will not enable public health workers to investigate any source of infection. In case of outbreaks the identification to the species level and molecular typing methods such as molecular fingerprinting would be necessary to provide evidence to follow the source of any infection. An outbreak of Streptococcus zooepidemicus caused by goat cheese produced from unpasteurized milk in a small dairy on a farm was reported. Streptococcus
zooepidemicus isolates from cheese, bulk milk, goats and also from seven patients with
severe invasive illness were typed using two different molecular methods. The investigation provided strong evidence that the source of the outbreak was the dairy products from the unpasteurized milk (Kuusi et al. 2006).
The two main species of the large colony streptococci Lancefield group C are
Streptococcus dysgalactiae subsp. equisimilis and the Streptococcus equi subsp. zooepidemicus. The Streptococcus equisimilis has been mainly detected in human
samples as a commensal or as a pathogen. While the other species, the Streptococcus
20
sporadic occurrence of its infections which is usually as a result of close contact with domestic animals, consumption of unpasteurized milk products and also with consumption of pork, this species is considered the most aggressive human pathogen among all members of the group C streptococci. When an increasing number of severe or even mild infections were detected in hospitals and clinical microbiology laboratories the application of molecular markers is necessary for precise characterization of the isolates. An article showed that detecting the restriction patterns of isolates in an outbreak investigation was a significantly useful way for the determination of the infection source and the right treatment application. Fifteen patients with a median age of 70 years were infected with the beta hemolytic zoonotic Streptococcus zooepidemicus. Serious infections with this species have been recorded more frequently in patients in over seventy or in neonates but rarely in young adults. In this outbreak five patients died while the others showed severe clinical signs of bacteremia, pneumonia, septic arthritis and also meningitis. The variation of severity indicates that some isolates might be actually more virulent than others. The molecular typing in this study was considered as an important contribution for the prevention of future infections when all the isolates displayed the same macrorestriction pattern by pulsed-field gel electrophoresis (Bordes-Benítez et al. 2006).
1.11.1. Random Amplification of Polymorphic DNA
The molecular markers are important tools used for the epidemiological investigations and variation. It is a detectable piece of DNA that can be used to produce a genomic or chromosomal map. This can be achieved by the various molecular techniques of biotechnology. Random amplified polymorphic DNA (RAPD) is one classical example of the techniques. In this method a short oligonucleotide primers are selected arbitrarily to amplify random sites on the genome (Kumarsen 2010).
The objective of the conventional PCR is the amplification of a specifically defined target sequence. The PCR technique is also employed in RAPD but the short primers used bind to a randomly distributed sites of the genome which vary among strains and this variation can be seen as patterns. The patterns may occur in a strain but not in the other (Satyanarayana 2008).
21
AP-PCR term is also used to describe the RAPD technique. Arbitrary amplification of polymorphic DNA sequences is a traditional molecular typing method that has been employed in many studies because of its advantages. It is relatively inexpensive and rapid and can be performed in most of the laboratories. In spite of its advantages RAPD technique always requires standardization in order to obtain clear reproducible results. This can be achieved by optimizing the PCR reaction contents and conditions and also the used visualization technique (Vogel et al. 1999).
1.12. Biofilm formation and its contribution to pathogenicity:
Another factor that may contribute to the disease development is the biofilm. Every passing day facing infections related to biofilm is becoming a bigger problem. The negative impact of biofilm can be seen in health and industrial sector as well. During infection the formation of biofilm layer prevents the contact of immune cells with the invading bacterial populations. It also helps the microorganisms to resist extreme heat, PH, UV and other environmental conditions. Inside the intestine, the biofilm formation on the epithelial surface protects the pathogens and allows them to thrive without any outside disturbance (Çiftçi 2005; Lindsay and Von Holy 2006).
According to some publications around 80% of all infections some of which are difficult to eradicate involve biofilms. Biofilms are formed as a sessile of bacterial communities. These bacterial cells are attached irreversibly to each other and embedded to a substratum. This is facilitated by the bacterial production of extracellular polymeric substances that make up the matrix that surrounds and contains the bacterial population. This structure allows the communication among bacteria by some chemotactic particles in a process called quorum sensing. Many factors contribute to the formation of biofilm. The availability of nutrients, surface adhesins, bacterial motility, chemostatics are examples of these factors (Hassan et al. 2011).
The presence of bacterial populations within a biofilm makes it remain exposed to antibiotics longer. This would enable the selection of more drug resistant populations and make the future application of treatment more and more difficult even with the high concentration (Di Bonaventura et al. 2004; Wilcox et al. 2001).
22
The bacterial biofilm layers formation is widely distributed in nature and it is the base for the development of various infectious diseases and resistance genes exchange among bacteria. The interventional and consistent techniques used in medical applications increase the occurrence rate of biofilm based infections. Gram positive and gram negative bacteria along with fungi are increasingly seen forming biofilm on various medical and instruments such as catheter, artificial heart valves and many other plastic tools and this contributes to the increasing hospital infections (Donlan 2002; Monzón et al. 2001).
1.12.1. Biofilm detection:
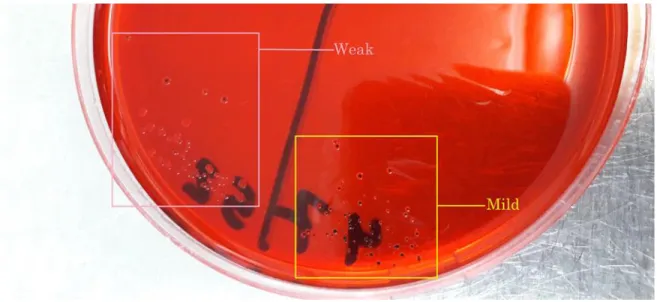
The detection of the microorganism’s ability to form biofilm can be assessed by several ways. Congo red agar is one of the rapid ways for the detection of biofilm in bacteria. Bacterial isolates are streak cultured on brain heart agar supplemented with sucrose and congo red dye. The formation of blackish colonies after incubation represents the strains with the ability to form biofilm (Cotter et al. 2009).
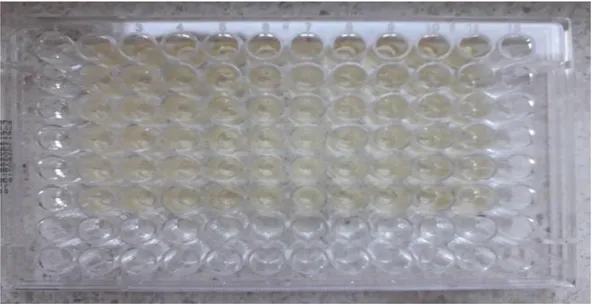
The degree of biofilm can be measured by using the optical density of the stained biofilm. Pure bacterial isolates are cultured in a sugar containing bacterial broth in a sterile micro plate wells. After incubation, the formed biofilm is washed and dried. After washing, crystal violet is then used for staining. The stained biofilm is then solubilized in a solution containing methanol and acetic acid which is then read in optical density reader (Moore 2009).
1.13. Antimicrobials and group C Streptococcus
In order to combat infectious diseases in horses, it is important to take protective measures and to prevent the spread of the disease without causing major losses and to apply the correct treatment methods. Turkey is well know with its high quality of animal production. Various antibacterial drugs are used to treat infectious diseases encountered in horses and other economically important animals. As of December 2017, numerous number of licensed antibiotic preparations in a form of solutions, suspensions, tablets, spray, ointment, drops, menstrual injection and eye-ear applications are available in the
23
country. The most importants among these preparations are the narrow and broad spectrum penicillins, aminoglycosides and sulfonamide (GKGM 2017).
In clinical microbiology laboratories testing the susceptibility of various bacterial isolates to antibiotics has become one of the main responsibilities. It is considered as important as the determination of the etiologic agents of infection. There is a real need for selecting a suitable antibiotic for the treatment of infections especially with the increasing resistance cases and also the increasing expenses of treatment. Identifying the pathogens is not enough for selecting the right theraputic agent. Some bacterial species such as
Strpetococcus pyogenes have predictable sensitivity to antimicrobials such as penicillin.
But various bacterial species known in microbiology have demonstrated resistance and this makes it difficult to predict the efficient antimicrobial that can be used for the treatment of a particular infection. The antimicrobial susceptibility test (AST) is also important for the evaluation of the activity of new and experimental antimicrobial compounds. Agar disc diffusion is one of the routinly used methods in most of the laboritories which allows the categorization of the susceptibility of various isolates to a wide range of antibiotics (Jenkins and Schuetz 2012).
According to several published studies equine beta-hemolytic streptococci are susceptible to the most of the commonly used antimicrobial agents including penicillin, ampicillin, amoxicillin and enrofloxacin. But records have shown resistance to gentamicin up to 14% of isolates, tetracycline up to 30% and higher resistance up to 95% was to sulphonamide. Human isolates also seem to be sensitive to erythromycin, vancomycin, benzylpenicilin, cephalosporin, rifampin and levofloxacin. In case of strangles, penicillin administration is usually recommended unless abscesses have already formed (Eyre et al. 2010; Jovanović et al. 2008; Pelkonen et al. 2013; Las Heras et al. 2002; Priestnall and Erles 2011).
24
2. MATERIALS AND METHOD
All the procedures throughout the study were performed under the biosafety cabinet class II in Microbiology laboratory of the Veterinary Faculty of Selcuk University.
2.1.The subculturing of Streptococci
All the 115 Streptococcus isolates had been purified and grown in brain heart broth and then stocked in a deep freeze temperature -80ºC. During the subculturing, the stock tubes were taken and kept in room temperature to melt. Samples were first cultured on the selective media of Streptococcus Edwards’s agar (Oxoid) to ensure the purity of the isolates. After that they were cultured on blood agar (LAB) for the colony observation and also in order to detect the hemolysis. All the cultures for isolation purpose in this study were usually incubated in 37°C in 5% CO2 containing atmosphere for 24-48 hours.
2.1.1. Blood agar base formulation (LAB)
Beef extract 10 g/L
Balanced peptone 10 g/L
Sodium chloride 5 g/L
Agar 12 g/L
Blood agar preparation
The amount of blood agar base used was weighted according to the producer (LAB). 37 gram is added to a liter of distilled water. It is mixed well and then autoclaved in 121°C for 15 minutes. The agar then is cooled down until it comes to 45°C. In that suitable temperature 50 ml of sterile sheep blood was added. The agar base and the blood are mixed well before pouring into sterile petri dishes. It is left to solidify and it had been kept in a cool place to avoid drying out until it was used.
2.1.2. The modified Edwards’s agar formulation (Oxoid)
Lab-lemco powder 10 g/L
Peptone 10 g/L
25
Sodium chloride 5 g/L
Crystal violet: 0.0013 g/L
Thallous sulphate: 0.33 g/L
Agar 15 g/L
Edwards’s agar preparation
The amount of Edwards agar base used was weighted according to the producer (oxoid). 41 gram was added to a liter of distilled water. It was mixed well and then autoclaved in 121°C for 15 minutes. Before adding the sterile sheep blood, the agar had been cooled to 45°C. 50 ml of sterile sheep blood was added and mixed well. It was then poured to sterile petri dishes and left to solidify.
2.2. The identification of group C Streptococcus
In order to encourage the growth of the streptococci optimum conditions were provided. The cultures were incubated in 5% CO2 aerobic (microaerophilic incubator) at 37°C for 24 to 48 hours. Some of the bacteria grew after 12 hours while some of them required 48 hours to form visible colonies.
2.2.1. Colony characterization
Before performing the biochemical identification the colony morphology was observed on blood agar before staining and observation under microscope. The colonies of the Lancefield C positive isolates on blood agar were observed prior to staining in order to initiate the identification. The colonies were water drops like transparent and small (1mm in diameter). The detection of the wide clear beta hemolysis zone around the streaking lines was the first characteristic to be observed in order to start the identification of this species.
2.2.2. Gram staining and microscopic observation
One drop of clean distilled water is mixed well with a loop full bacterial colony on a slide. The slide is kept to air dry for few minutes. The smear is then fixed by holding the slide for few seconds on the flame. After fixation, crystal violet is added and waited for 3 minutes. It is then washed with fluent water. Lugol’s Iodine Solution is then added and washed after waiting for one minute. The slide is then washed with 95% ethanol and
26
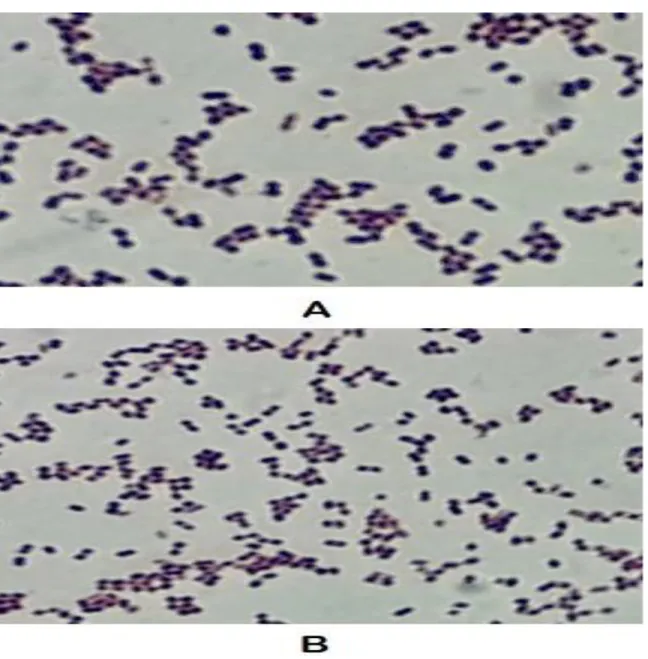
kept for about a minute before washing with water. In the last step, basic fuchsin dye is added to the slide and washed after 30 seconds. The slide is then dried using tissue paper. One drop of immersion oil is usually added on the smear before the microscopic examination with 100x. The expected bacterial morphology of the isolates in this study was gram positive slightly oval and some larger round cocci in chains.
2.2.3. Biochemical identification Catalase test
This identification test is used for the detection of the catalase enzyme production by the tested organism. The presence of this enzyme means the ability of the organism to detoxify the toxic radicals such as Hydrogen peroxide. In this test one colony is placed on a clean slide. One drop of H2O2 is added. The clear bubbles formation and the release of gas is an indication that the organism is positive. In this study Staphylococcus aureus was used as a positive control (Leboffe and Pierce 2012). The strains used in this study were expected to be catalase negative.
Figure 2.1. The breakdown of hydrogen peroxide to oxygen and water by the catalase
activity.
Figure 2.2. Staphylococcus is on the left, Enterococcus is on the right. Catalase slide
27 Sugar fermentation test
The differentiation of the group C streptococci members is done based on the ability of the strains to ferment various sugars. In this study 3 sugars were used for the primary identification. These are lactose, sorbitol and trehalose.
Carbohydrate consumption broth formulation
Nutrient broth 1.5 gram/Liter
Sugar (lactose or sorbitol or trehalose) 1 gram/Liter
Bromocresol purple indicator 0.001 gram/Liter
Final pH (at 25°C) 6.8
1.5 g of nutrient broth was dissolved in 1 liter of distilled water. After adding the indicator and after well mixing the broth was autoclaved in 121°C for 15 minutes. A single sugar solution is sterilized by using a 0.2 microns filter. After adding the sugar to the sterile broth, 5 ml was distributed into each sterile glass tube. After inoculation the tubes were incubated in 5% CO2 37°C for 24 hours. The change of color from purple to yellow after 24 hours incubation represents the positive result. Streptococcus
zooepidemicus ATCC 35246 was used as positive control along with Escherichia coli.
The negative control used was sterile sugar broth.
2.2.4. Lancefield grouping latex test
In this study, 115 bacterial isolates that had been previously identified as
Streptococcus based on their colony morphology, microscopic observation and catalase
testing were tested using the strep PRO grouping kit. This test was performed according to the producer’s (Hardy diagnostics) protocol. The kit used consists of 3 extraction reagents and six colored coded reagents represent the streptococcal groups A, B, C, D, F, and G. One drop of reagent one is mixed with one colony in a clean test tube. One drop of reagent two is then added and mixed well. Five drops of reagent three is the added and again mixed well. Each group is tested by mixing one drop with one drop of the extracted antigen. The agglutination appears immediately which is an indication of the positive result. The isolates selection in this study was based on this test. Only Streptococci that gave positive C group were chosen. After the determination of the Streptococcus strains