1
ELUCIDATING IMMUNOMODULATORY EFFECTS OF
TELOMERIC REPEAT MIMICKING SYNTHETIC A151
OLIGODEOXYNUCLEOTIDE ON IMMUNE CELL
TRANSCRIPTOME
A DISSERTATION SUBMITTED TO
THE GRADUATE SCHOOL OF ENGINEERING AND SCIENCE
OF BILKENT UNIVERSITY
IN PARTIAL FULFILLMENT OF THE REQUIREMENTS
FOR THE DEGREE OF
DOCTOR OF PHILOSOPHY
IN MOLECULAR BIOLOGY AND GENETICS
by
Volkan Yazar
i
ELUCIDATING IMMUNOMODULATORY EFFECTS OF TELOMERIC REPEAT
MIMICKING SYNTHETIC A151 OLIGODEOXYNUCLEOTIDE ON IMMUNE
CELL TRANSCRIPTOME
by Volkan Yazar September, 2019
We certify that we have read this dissertation and that in our opinion it is fully adequate, in scope and in quality, as a thesis for the degree of Doctor of Philosophy.
İhsan Gürsel (Advisor)
Can Alkan (Co-advisor)
Tolga Can
Işık Yuluğ
Mayda Gürsel
Ali Osmay Güre Approved for the Graduate School of Engineering and Science
Ezhan Karasan
ii
ABSTRACT
ELUCIDATING IMMUNOMODULATORY EFFECTS OF TELOMERIC REPEAT
MIMICKING SYNTHETIC A151 OLIGODEOXYNUCLEOTIDE ON IMMUNE
CELL TRANSCRIPTOME
Volkan Yazar
Ph.D. in Molecular Biology and Genetics Advisor: İhsan Gürsel
Co-advisor: Can Alkan September, 2019
Recent evidence revealed that DNA is beyond just the blueprint of life; it is also involved in immunomodulation. Unmethylated Cytosine-phosphate-Guanine (CpG) motifs of prokaryotic DNA stimulate immune response by interacting with Toll-like receptor 9 (TLR9). This interaction is mimicked using synthetic oligodeoxynucleotides (ODN) bearing similar DNA motifs to boost vaccine-driven immune response in human. Conversely, mammalian telomeric ends expressing TTAGGG repeats suppress immune response and contribute to fine-tuning of delicate immune balance. In this respect, suppressive ODN A151 with such G-rich telomeric repeats has proven useful in downregulating immune response; an overly active immune response is just as harmful to the host, as in the case of autoimmune disorders. Both CpG ODN and A151 are currently under preclinical/clinical trials with the aim of averting or medically treating a wide range of conditions from cancer to infectious disease or from autoimmune to autoinflammatory conditions. Contrary to CpG ODN, A151 literature is very limited and its modus operandi at gene level remains more of a mystery. Additionally, the degree, duration and breath of A151-induced alterations in immune transcriptome appear partially understood. Given the medical potential A151 holds for immunosuppressive therapy in human as a “self-molecule”, understanding the underlying molecular mechanisms via which A151 operates is invaluable. Toward this end, we attempted to uncover the unidentified features lying behind A151 ODNs immunosuppressive effects on immune cell transcriptome using a combined analysis approach of microarray data in this thesis. We demonstrated for the first time that A151 ODN deprives the cells energy by ceasing cellular uptake of fundamental molecules into the immune cells after derailing the entire intracellular trafficking. Putting it another way, A151 does not directly act on immune system cells but actually suffocates the cells by messing with intracellular trafficking, thereby blocking cellular uptake of fundamental molecules like glucose and glutamine. As such, immune suppression is just an indirect consequence
iii
of this larger cellular chaos. Our results indicated that this phenomenon occurs independent of CpG ODN stimulation of the cells and in a timely manner. Most, if not all, regulators of intracellular trafficking, vesicle signaling, and membrane protein transportation were found downregulated after incubation of cells with A151 at a physiologically relevant concentration, as well, implying full-blown entry to this intracellular turmoil at cellular level. The A151 effect on immune transcriptome was not just restricted to setting off a chaos for intracellular dynamics; novel long non-coding RNAs (lncRNAs) with immunometabolic activities were identified within the scope of this study among elements potentially regulated by A151, such as Lncpint, Malat1 and H2-T10 just to name a few. The involvement of lncRNAs in immune regulation is a well-documented phenomenon. Finally, our data showed that as an epiphenomenon of the intracellular turmoil mentioned above A151 has a deep impact in immune cells on mTOR network, the cardinal network of cellular energetics, growth, proliferation, and survival. A major shift in expression profile of relevant genes, i.e. downregulation of many activators of mTOR signaling along with core mTOR components, was validated on the benchtop after different layers of experimental validation using a wide range of marker genes and functional assays, reflecting A151’s ability to vastly shape dynamics of metabolism in favor of a metabolically inert state in macrophages and in B-cells. This knowledge will expand the breadth of A151 therapy in the clinics.
Keywords: Innate immunity, CpG ODN, suppressive ODN A151, transcriptome analysis, microarray,
iv
ÖZET
TELOMERİK DNA BENZERİ SENTETİK A151 OLİGODEOKSİNÜKLEOTİD’LERİN
BAĞIŞIKLIK HÜCRE TRANSKRİPTOMU ÜZERİNE BAĞIŞIKLIK DÜZENLEYİCİ
ETKİLERİNİN AYDINLATILMASI
Volkan Yazar
Moleküler Biyoloji ve Genetik, Doktora Tezi Tez Danışmanı: Prof. Dr. İhsan Gürsel Tez Eş Danışmanı: Yrd. Doç. Dr. Can Alkan
Eylül, 2019
Yapılan son çalışmalar DNA’nın, yaşamın mavi baskısı olmasının ötesinde bağışıklık sistemini düzenleyici etkisini göstermiştir. Prokaryotik DNA’ya ait metil grubu içermeyen Sitozin-fosfat-Guanin (CpG) motifleri Toll-benzeri reseptör 9 (TLR9) ile etkileşerek bağışıklığı uyarır. Bu etkileşim, insanda aşılamanın kazandırdığı bağışıklığı artırmak amacıyla benzer DNA motifleri ihtiva eden sentetik oligodeoksinükleotidler (ODN) kullanılarak hayata geçirilmektedir. Öte yandan, tekrarlayan TTAGGG dizini içeren memeli kromozomlarının telomerik uçları bağışıklığı baskılayabilmekte ve hassas bağışıklık dengesinin ayarlanmasında katkı sağlamaktadır. Bu bağlamda, esasen G-zengini telomer dizini olan baskılayıcı ODN A151’in bağışıklığı baskılamada etkili olduğu kanıtlamıştır; otoimmün hastalıklarda olduğu gibi, aşırı boyutta etkin bir bağışıklık cevabı da kişiye zarar vermektedir. CpG ODN ve A151 hâlihazırda klinik faz/klinik öncesi test kapsamlarında kanser ve bulaşıcı hastalıklardan otoimmün ve otoenflammatuvar hastalıklara kadar birçok tıbbi bozukluğun tedavisi için denenmektedir. CpG ODN’nin aksine, A151 literatürü çok sınırlıdır ve gen seviyesinde çalışma prensibi bilinmemektedir. Buna ek olarak, A151’in bağışıklık transkriptomunda yaptığı değişikliklerin boyutu ve süresi tam anlamıyla aydınlatılamamıştır. A151’in hücreye ait “öz” bir molekül olarak tıbbi anlamda taşıdığı potansiyel göz önüne alındığında, A151’e ait işleyiş mekanizmasının aydınlatılması önem arz etmektedir. Bu amaçla, A151’in bağışıklık hücre transkriptomu üzerindeki bağışıklık düzenleyici etkilerinin anlaşılması adına birleşik mikroarray veri analizi tekniği kullanılarak bu tez çalışması hazırlanmıştır. A151’in hücre içi trafiği engellemesinin akabinde enerji zengini moleküllerin hücre içine alımını baskılamak suretiyle hücreleri enerjiden mahrum bıraktığını göstermiş olduk. Başka bir deyişle, A151 bağışıklık hücrelerini doğrudan etkilememektedir, bunun yerine hücre içi trafiği engelleyerek glikoz, glutamin gibi moleküllerin hücreye alınıp içeride yıkımını yavaşlatır ve böylece hücre metabolizmasını baskılar. Bağışıklığın baskılanması ise, bizim sonuçlarımıza göre, bu büyük etkinin dolaylı bir sonucudur. Yaptığımız çalışmalar bu gözlemin bağışıklık hücrelerin CpG ODN
v
ile etkinleştirilmesinden bağımsız ve zamana bağlı bir şekilde ortaya çıktığını göstermiştir. Hücrelerin fizyolojik olarak uygun miktardaki A151 ile inkübasyonu sonucu hücre içi trafik, kesecik sinyalizasyonu ve zar proteinlerinin taşınmasında görevli birçok denetleyici genin baskılandığını ispatladık. Ancak, A151’in bağışıklık hücre transkriptomu üzerindeki baskılayıcı etkilerinin bununla da sınırlı kalmadığını gördük; Lncpint, Malat1 ve H2-T10 gibi uzun protein kodlamayan RNA (lncRNA)’ların A151 tarafından denetlenen elementler arasında olduğunu bu çalışma kapsamında göstermiş olduk. lncRNA’ların bağışıklık denetlemesiyle olan ilişkisi hâlihazırda bilinmektedir. Son olarak, sonuçlarımız A151’in bağışıklık hücrelerindeki enerji döngüsü, büyüme, bölünme ve hayatta kalma işlevlerinden sorumlu birincil yolak olan mTOR sinyal yolağına büyük bir etkisi olduğunu ima etmiştir. mTOR yolağını etkinleştiren ve bu yolağın temel parçası olan birçok genin ifade miktarındaki değişimler farklı birtakım belirteç (marker) gen ve fonksiyonel incelemeler kullanılmak suretiyle gösterilmiş olup, bu da A151’in makrofajlarda ve B-hücrelerinde metabolik dinamikleri şekillendirmede hayati rolünü gözler önüne sermektedir. Bu bulgular A151 ODN’nin klinik uygulamalarının genişletilmesine ve hayata geçirilmesine katkılar sağlayacak niteliktedir.
Anahtar kelimeler: Doğal bağışıklık, CpG ODN, baskılayıcı ODN A151, transkriptom analizi,
vi
vii
Acknowledgements
First and foremost I want to thank my academic father and my advisor Prof. İhsan Gürsel for every single thing he has done for me even before this Ph.D. opportunity emerged, including his guidance through my graduate education abroad, his time for our endless scientific and non-scientific discussions that helped shape my career and basically my life, his constant support throughout almost all the decisions I have made so far, which are just few of many things I should be thanking him for. I would like to express my eternal gratitude to him for being who he really is, and in fact, consider myself very lucky to have had such an encounter at this stage of my career. I recognize that his limitless patience, field knowledge, and scientific guidance made this Ph.D. work possible. I am forever in his debt. I, also, acknowledge that my co-advisor Assist. Prof. Can Alkan of Computer Science helped me in gaining an additional layer of perspective to coding, data structures, algorithms, and big data analysis. He did facilitate my personal growth in the field of Computer Engineering both theoretically and practically. Also, all the Computer Engineering courses I have followed on campus were made possible through the heads of the CTIS Department, Bilkent; Dr. Erkan Uçar and Dr. Serpil Tın. I cannot thank them enough for that.
It would be unfair on my part if I let my office mates in Computer Science go unaccounted for. I personally thank EA507 crew for embracing me as one of their own and for helping me in developing a career in Computer Science, not just in Bioinformatics. My time at Bilkent was made enjoyable in large because of them.
In regard to TTC meetings, I would also like to thank my tracking committee members; Assist. Prof. Özgür Şahin (of Molecular Biology and Genetics) and Prof. Tolga Can (of Computer Engineering) for their precious time, as well as insightful questions that fine-tuned the directionality of my thesis study.
Moreover, I gratefully acknowledge the funding sources that made my Ph.D. work possible. I was funded by Bilkent University Department of Molecular Biology and Genetics, as well as Department of Computer Science, for the entire duration of my studies. This work was partially supported by Scientific and Technological Research Council of Turkey (TUBITAK) [grant number 115S492 to I.G.], [grant number 115S837 to I.G.] and Ministry of Development [grant name UMRAM-ASI to I.G.].
viii
Lastly but not least importantly, I would like to thank my big family for all their logistic and psychological support and for creating more stimulating less stressful home environment throughout the course of this Ph.D. work. I honestly express my deepest love to my mom, dad, and brother Gönül, Uluer, and Umut Yazar for their everlasting support and faith in me. They raised me with a love of science and supported me in all my pursuits. My grandparent Muammer Yazar who passed away years ago has always led me to achieve more than I could have ever dreamed of and will hopefully do so spiritually for the rest of my life. Thank you.
ix
Contents
ABSTRACT………...ii ÖZET……….………..………..…iv Acknowledgements………..……….…..vii Contents………..………...ix Lists of Figures………..………..…xiii Lists of Tables………..………..xv Abbreviations……….……….…xvi 1. Introduction……….1 1.1 Immune System………...…11.2 Innate Immune System………..………….……..2
1.3 Pattern Recognition Receptors……….……….……..3
1.4 Toll-like Receptors……….……….…..3
1.4.1 Cell Surface Toll-like Receptors……….……….……..5
1.4.1.1 TLR1, TLR2, TLR6………...…..5
1.4.1.2 TLR4……….……..…6
1.4.1.3 TLR5……….….….…6
1.4.1.4 TLR11……….6
1.4.2 Intracellular Toll-like Receptors……….…….…….….7
1.4.2.1 TLR3……….……….…….…8
1.4.2.2 TLR7 and TLR8……….……….…….….8
1.4.2.3 TLR9……….……….…….…9
1.4.2.4 Trafficking of Intracellular TLRs……….………..…..10
1.4.2.5 Endosomal Cleavage of Intracellular TLRs…….………….…….……….….….11
1.5 TLR Signaling……….………..……12
1.5.1 MyD88-dependent Pathway……….……..….13
1.5.2 TRIF-dependent Pathway………...……14
x 1.6.1 Immunostimulatory CpG ODN……….….….15 1.6.2 Immunosuppressive ODN………..……16 1.7 p53……….21 1.8 GAPDH……….………..……….22 1.9 Apoptosis……….………..…..…23 1.10 Cellular Senescence……….………..….…..23
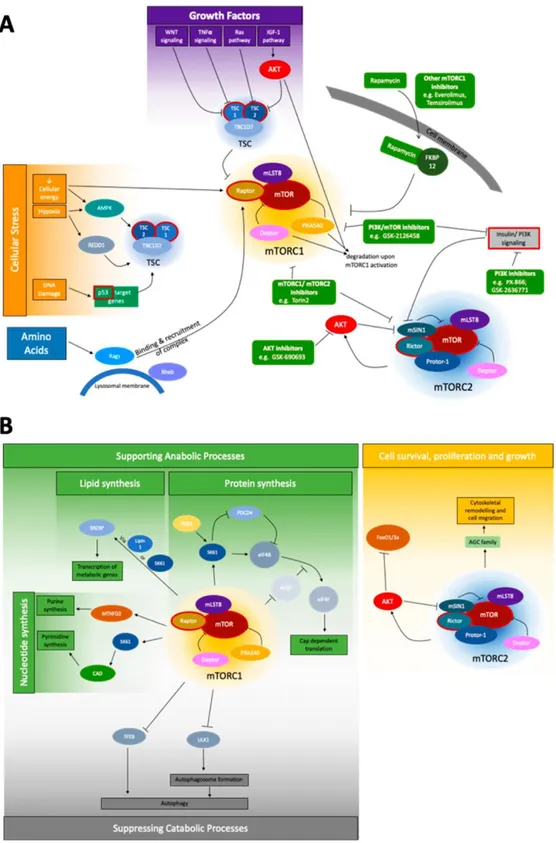
1.11 mTOR Signaling Pathway……….………..…....26
1.12 Aim and Strategy………..……32
2. Materials and Methods………..….35
2.1 Microarray Dataset Collection………35
2.2 Data Retrieval from mAdb and Experimental Design of Microarray Analyses………….35
2.3 Differential Expression Analysis……….………38
2.3.1 BRB ArrayTools………...…..38
2.3.2 Data Pre-processing………...…38
2.3.3 Visualization of Filtered Data………..….…39
2.3.4 Clustering of Filtered Data………..….….41
2.3.5 Pairwise and Ternary Comparisons……….….……..41
2.3.6 Meta-Analyses……….….….…43
2.3.7 Time-series Analyses………..….….45
2.3.8 Analyses of All Groups Combined at Individual Time Points……….…...45
2.3.9 Analyses of Each Time Point per Group……….……..…..46
2.3.10 GO Enrichment Analysis for Immune Relevance………...……47
2.3.11 Utility-based Comparison of Gene Lists………47
2.4 Identification of Long Non-coding RNAs in the Gene List……….……..49
2.5 Ingenuity Pathway Analysis………..……..50
2.6 Identification of TP53 and its Associates within A151 Treatment Groups…………..……51
2.7 Identification of Expression Profiles of Pro-Apoptotic and Pro-Survival Genes within A151 Treatment Groups………..………….……….51
2.8 Identification of Senescence Marker Genes within A151 Treatment Groups…………..52
2.9 Identification of mTOR Network Genes within A151 Treatment Groups……….…52
2.10 Code-based Comparison of Gene Lists………..……….………..52
2.11 STRING (v10.5) Analysis……….………..…………53
2.12 Reagents………..………...….….54
2.13 Bone Marrow Derived Macrophage (BMDM) Generation………..54
xi
2.15 Total RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
(qRT-PCR)………..……….…..55
2.16 Intracellular Staining for mTOR Phosphorylation…………..………...…55
2.17 Immunoblotting analysis……….……….………..55
2.18 Metabolic Assays……….……….………56
2.19 Statistical Analysis……….……….……….56
3. Results………..………….……57
3.1 Differential Expression Analysis……….…57
3.1.1 Data Quality Assessment……….………57
3.1.1.1 2D and 3D Scatterplots……….…….57
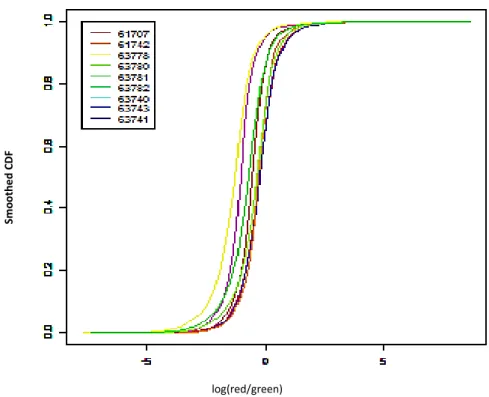
3.1.1.2 Smoothed Cumulative Distribution Function (CDF) Plot……….…..58
3.1.1.3 Boxplot of Gene Expression………59
3.1.1.4 Histogram of Expression Data………..………..…….61
3.1.1.5 Pairwise Correlation Plot……….……….….….62
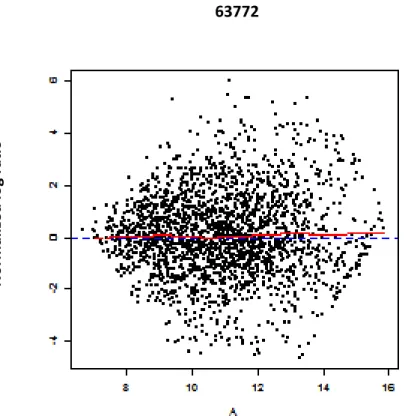
3.1.1.6 Dual Channel M vs. A (MA) Plot………..………..….63
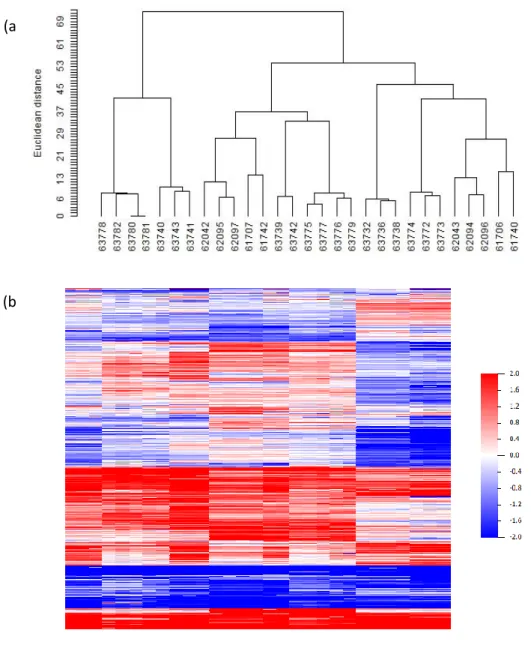
3.1.2 Clustering Analysis………..……….……..64
3.1.3 Class Comparison………..……….….66
3.1.3.1 Pairwise and Ternary Comparisons………..……….…..66
3.1.3.2 Meta-Analyses………..……….….70
3.1.3.3 Time-series Analyses………...………..……….…..71
3.1.3.4 Analyses of Each Time Point per Group………...…….74
3.2 GO Enrichment Analysis of Immune Relevance………..……….…….76
3.3 Identification of Long Non-Coding RNAs on the Lists……….77
3.4 Ingenuity Pathway Analysis for Immune Relevance……….……….……….……..77
3.5 Identification of TP53 and its Associates within A151 Treatment Groups………..81
3.6 Identification of Expression Profiles of Pro-Apoptotic and Pro-Survival Genes within A151 Treatment Groups……….………..….83
3.7 Identification of Senescence Marker Genes within A151 Treatment Groups…………..84
3.8 Identification of mTOR Network Genes within A151 Treatment Groups…….……..……87
3.9 Ingenuity Pathway Analysis for mTOR signaling………….………..……88
3.10 Physical Validation of A151 Effect on mTOR Signaling……….………..……..90
3.10.1 qRT-PCR Validation of Expression Levels of mTOR Network Genes……….90
3.10.2 Flow Cytometry Validation of mTOR Activity……….……….….…91
3.10.3 Western Blot Validation of mTOR Activity………..…..92
xii
3.11 A Comprehensive Elucidation of the A151-based Immune Suppression………95
3.12 Physical Validation of A151 Effect on Immune Cells……….………..……99
4. Discussion………...…...100
4.1 Differential Expression Analysis……….………..….102
4.1.1 Data Quality Assessment……….………..….102
4.1.2 Clustering Analysis……….………..……104
4.1.3 Class Comparison……….……..…..105
4.2 GO Enrichment Analysis for Immune Relevance……….…..……110
4.3 Identification of Long Non-Coding RNAs on the Gene Lists………...……110
4.4 Ingenuity Pathway Analysis for Initial Insight into the Findings ……….…111
4.5 Identification of TP53 and its Associates within A151 Treatment Groups………...…..113
4.6 Identification of Expression Profiles of Pro-Apoptotic and Pro-Survival Genes within A151 Treatment Groups………..………114
4.7 Physical Validation of A151 Effect on Apoptosis and Senescence………..……..115
4.8 Identification of mTOR Network Genes within A151 Treatment Groups………….…...115
4.9 Ingenuity Pathway Analysis for mTOR signaling……….….……..116
4.10 Physical Validation of A151 Effect on mTOR Signaling……….….…….116
4.11 A Comprehensive Elucidation of the A151-based Immune Suppression………….…..118
5. References……….…….121
6. Appendices……….……135
7. Curriculum Vitae and Publications……….…..145
xiii
List of Figures
Figure 1.1. PAMP recognition by cell-surface TLRs (Adapted from [3])………..………7
Figure 1.2. PAMP recognition by intracellular TLRs and the role of UNC93B1 in their trafficking from ER to endolysosome (Adapted from: [3])……….………11
Figure 1.3. TIR domain containing adaptors in TLR signaling (Adapted from [27])……….………...12
Figure 1.4. MyD88 and TRIF dependent pathways (Adapted from [15])……….……….………….13
Figure 1.5. D- (CpG A) and K-type (CpG B) ODN induced signaling pathways and their intracellular localization (Adapted from [82])……….……….………..16
Figure 1.6. Diverse effects of suppressive ODN A151 on cellular elements of the immune system (Adapted from [86])……….…….………..19
Figure 1.7. Major pathways of replicative and stress-induced premature senescence (Adapted from [122])……….………25
Figure 1.8. The mTOR signaling network (Adapted from [182])………28
Figure 3.1. 2D and 3D scatterplots for data quality assessment.……….58
Figure 3.2. Smoothed CDF plot for data quality assessment………..59
Figure 3.3. Boxplots of gene expression for data quality assessment………..60
Figure 3.4. Histograms for data quality assessment………..………..61
Figure 3.5. Pairwise Correlation Plot for data quality assessment………..………..62
Figure 3.6. MA Plot for data quality assessment………..………..63
Figure 3.7. Visualization of clustering analysis of the selected data sets……….……….65
Figure 3.8. HeatMap of significant genes selected from TC3 analysis at time point 8 hours…………..…67
Figure 3.9. Parallel coordinate plot of the outcome of TC3 analysis at time point 8 hours……..…………68
Figure 3.10. Benchmarking of two distinct methodologies for microarray data analysis………..…………71
Figure 3.11. HeatMap of significant genes selected from time-series analysis of untreated sample…72 Figure 3.12. Interrupted time-series regressions (expression profiles of three significant genes selected from time-series analysis of untreated data sets)……….73
Figure 3.13. GO Enrichment Analysis for a selected gene list………76
Figure 3.14. Ingenuity Pathway Analysis (IPA) network 1.1……….………78
Figure 3.15. IPA network 1.2……….80
Figure 3.16. Molecular mechanisms of cellular senescence [142]……….………85
Figure 3.17. IPA network 2.1……….89
xiv
Figure 3.19. Flow cytometry and Immunoblotting results of mTOR……….93
Figure 3.20. Metabolic profiling of A151-treated immune cells by SeaHorse Analyzer……….….94
Figure 3.21. STRING network for the final list of genes……….……….96
Figure 3.22. A proposed mechanism of action for A151-based immune suppression……….……98
xv
List of Tables
Table 1.1. Chromosomal localizations of TLRs [3, 11]……….4
Table 1.2. TLRs and their agonists [10, 13]……….5
Table 1.3. Cellular localization of endosomal TLRs [50 ,51]……….………..8
Table 1.4. The effects of suppressive ODN on CpG-induced immune activation……….17
Table 1.5. The effect of Suppressive ODN on IFNγ production induced by several immune. activators………..………..18
Table 1.6. mTOR pathway-associated genes (Adapted from [182])………..29
Table 2.1: Number of arrays used for each treatment group per time point………..………..36
Table 2.2: Main comparisons performed for each time point 1 hour, 2 hours, 4 hours, and 8 hours separately………..……….36
Table 2.3: Another round of experiments including different pairwise comparisons………..37
Table 2.4: Design for gene list comparisons of PC……….………48
Table 2.5: Design for gene list comparisons of TC……….………49
Table 3.1: Complete set of analysis results obtained from “Between Groups of Arrays” tool of BRB ArrayTools……….………….69
Table 3.2: Complete set of analysis results obtained from “Time-course Analysis” tool of BRB ArrayTools...74
Table 3.3: Complete set of analysis results obtained from “ANOVA on log intensities for two-color arrays” tool of BRB ArrayTools……….………75
Table 3.4: A complete list of TP53-associated genes that are differentially expressed with statistical significance after A151 treatment available in the gene lists indicated……….…81
Table 3.5: Two gene lists that show A151-induced up- and downregulation of gene expression (of pro-apoptotic genes and of pro-survival genes, respectively)………..83
Table 3.6: A selected list of senescence markers found in our A151-associated gene lists and their expression pattern after A151 treatment……….………..86
Table 3.7: A selected list of mTOR network genes found in our A151-associated gene lists and their expression pattern after A151 treatment……….…..88
xvi
Abbreviations
A151 Suppressive oligodeoxynucleotide
Ab Antibody
ANOVA Analysis of Variance
APC Antigen presenting cell
BCR B-cell receptor
BMDMs Bone marrow derived macrophages
bp Base pairs
BPG Biphosphoglycerate
CARD Caspase activation and recruitment domain
CD Cluster of differentiation
CDF Cumulative Density Function
cDNA Complementary Deoxyribonucleic Acid
CIA Collagen-induced arthritis
CLR C-type lectin receptors
CpG Unmethylated cytosine-guanosine motif
CXCL CXC-chemokine ligand
Cy3 Cyanide 3 dyes
Cy5 Cyanide 5 dyes
DC Dendritic cell
DNA Deoxyribonucleic acid
dsRNA Double-stranded RNA
ER Endoplasmic reticulum
FASL Fas ligand
FcR Fc Receptor
FDR False discovery rate
GAPDH Glyceraldehyde-3-phosphate dehydrogenase
GGIN Gene-gene interaction network
GO Gene Ontology
GPI Glycosylphosphatidylinositol
IFN Interferon
xvii
IL Interleukin
IPA Ingenuity Pathway Analysis
IP-10 Interferon gamma-induced protein 10
IRAK IL-1 receptor-associated kinase
IRBP Interphotoreceptor retinoid-binding protein
IRF3 Interferon-regulatory factor 3
KO Knock out
ICAM-1 Intercellular Adhesion Molecule 1
IFI16 Gamma-interferon-inducible protein
IκK Inhibitor kappa B kinase
INH ODN Inhibitory oligonucleotides
LLR Leucine-rich repeats
LBP LPS binding protein
LPS Lipopolysaccharide
lncRNA Long non-coding RNA
MA Log-ratios average intensity plot
mAb Monoclonal antibody
mAdb the Microarray Database
MAP Mitogen-activated protein
MAPK Mitogen-activated protein kinase
MD Myeloid differentiation protein
MDS Multidimensional scaling
MHC Major Histocompatibility Complex
MIP Macrophage Inflammatory Protein
MS ODN Microsatellite DNA mimicking ODNs
mTOR Mechanistic target of rapamycin
MyD-88 Myeloid differentiation primary response gene (88)
NZB/W New Zealand Black/White
NF-κB Nuclear factor-kappa B
NK Natural killer
NKG2D Natural-killer group 2, member D receptor
NLR NOD-like receptor
NOD Nucleotide-binding oligomerization domain
ODN Oligodeoxynucleotide
xviii
PC Pairwise comparison
PCA Principle Component Analysis
PCR Polymerase chain reaction
PCP Pairwise correlation plot
pDC Plasmacytoid dendritic cells
PECs Mouse peritoneal exudate cells
PGN Peptidoglycan
pIC (polyIC) Polyriboinosinic polyribocytidylic acid
PPIN Protein-protein interaction network
PRR Pattern Recognition Receptors
pRB Retinoblastoma protein
p16 Protein 16
p21 Protein 21
p53 Protein 53
RIG Retinoic acid-inducible gene
RLR RIG-like receptor
RNA Ribonucleic acid
RVS Respiratory syncytial virus
RT-PCR Reverse transcriptase PCR
SAM Statistical Analysis of Microarray
SOCS Suppressor of cytokine signaling
STAT Signal transducer and activator of transcription
ssDNA Single stranded DNA
ssRNA Single-stranded RNA
TC Ternary comparison
TCR T-cell receptor
TH T-helper
TIR Toll/IL-1 receptor
TIRAP Toll/IL1 receptor-associated protein
TLR Toll-like Receptor
TNF Tumor Necrosis Factor
TRAM TRIF-related adaptor molecules
1
Chapter 1
Introduction
1.1 Immune SystemThe immune system is a network of organs and processes of the body which protect the host from pathogens and insults of various kinds. Considering the evolutionary tree of life, all living species have been adapted to defend themselves against any danger; primitive organisms such as bacteria and algae have less developed mechanisms for this purpose while higher eukaryotes have been engineered with more elaborate defense instruments. Lower level eukaryotes like invertebrates and less sophisticated plants use the fundamental branch of immunity called innate immune system while higher level eukaryotes such as mammals have an extra layer of protection named adaptive immune system, as well [1, 2].
Innate immune system is known as a front line of defense within the scope of immunity; it is quick and extremely effective in dealing with intruders. The molecular mechanisms involved are, as the name implies, innately acquired and subject to neither alteration nor modification. Even though innate immune system is not specific to a particular antigen, it handles different pathogens differently (in a way, specifically). This type of immune system “senses” different pathogens using so-called “sensors” named pattern-recognition receptors (PRRs). These PRRs are exceptionally conserved and have been specialized to detect pathogen-associated molecular patterns (PAMPs) [3, 4]. Nevertheless, the well-established doctrine in the field that innate immunity has no memory to remember the same pathogen at a possible next encounter has been recently challenged with the evidence that innate immune cells did and can maintain a memory for a higher magnitude response for next encounter with the same pathogen within the context of “trained memory” or “innate immune memory” [5, 6].
Adaptive immune system, also known as acquired immune system, is another branch of mammalian immune system that uses specific antigens or even epitopes to strategically mount an immune response as a second line of defense. Clonal expansion of antigen-specific lymphocytes, or B- and T-cells, takes longer than the immediate response mounted by innate immunity does, delaying
2
response time of adaptive immunity to pathogens. Receptor repertoires of these highly specific cells are vast, thanks to the random rearrangement of V, D, and J segments in the genome. The distinguishing characteristic of adaptive immunity is its ability to develop protection against re-infection called immunologic memory [1]. The underlying mechanism of innate immune system relies upon the activation of “cellular” response composed of CD4+ T helper cells (Th1 and Th2), and cytotoxic T cells (CTLs), and subsequent revival of “humoral” response as antigen-specific antibody production by B cells [3]. Non-self antigens that interact with a special type of receptors on cell surface called major histocompatibility complex class I (MHC-I) are recognized by CTLs and this recognition results in extermination of the infected cell, including cancer cells. Antigen presenting cells (APCs) like dendritic cells maintain antigen presentation by presenting these non-self antigens on their MCH-II cell surface receptors to T cells. Priming of these T cells needs an extra layer of stimulation, which is done by upregulation of co-stimulatory molecules to be secreted to the site of interaction. Subtypes of T cells are generated only by activated T cells and these highly specific T cell subtypes secrete particular cytokine molecules that can aid B cells in sensing non-self antigen and in producing customized antibodies [7].
1.2 Innate Immune System
As the first line of defense, innate immune system is composed chiefly of complement system, phagocytes, and APCs. Reacting rapidly, innate immunity acts to hamper spreading of infection at the onset of invasion. Neutralization of the insult is followed by antigen presentation of APCs to adaptive immune system cells for them to transform into antigen specific effector cells. These APCs such as macrophages and especially dendritic cells play a pivotal role in protecting the host from pathogens and the vast majority of foreign entities; they digest pathogen-specific molecules to sizeable amounts and the resulting epitopes are presented to B- and T-cells to initiate antigen-specific immunity [8, 9].
A substantial versatility in host defense mechanisms against a wide range of pathogens showing massive molecular diversity per se was crafted to almost perfection by evolution over the time upon numerous encounters in a struggle called survival. In this arms race a particularly important innate immune mechanism named phagocytosis provides host immunity with a salient advantage of directly engulfing and degrading pathogens into monomers after a sequential order of events. Reactions as such, given by a healthy immune system against invaders, are meant to direct the host to rebalance a disrupted homeostasis or physiological state. To put it another way, the degree of this immune response given during infection to reconstitute homeostasis is balanced in healthy systems. It is this balance that reassures the continual existence and proper functioning of the organism itself.
3 1.3 Pattern Recognition Receptors (PPRs)
Specific conserved patterns of microbial world, or signature pathogenic components, are recognized by innate immunity using PPRs. Most commonly known PPRs are Toll-like receptors (TLRs), which are described below neatly. Some TLRs act as endosomal nucleic acid sensors due to their ligand-specific spatial localization inside the cell. They detect unusual molecular patterns of pathogens like unmethylated CpG motifs or dinucleotides and cause strong innate immune activation [11]. They are so effective in doing this that nucleic acid based adjuvants were taken to preclinical trials and suggested to be included safely within vaccine formulations [12, 13].
Other examples of PPRs are retinoic acid-inducible gene I (RIG-I) -like receptors (RLRs), nucleotide-binding oligomerization domain (NOD) like receptors (NLR) and C-type lectin receptors (CLR) together with cytosolic DNA and RNA sensors. The microbial ligands of PPRs are called pathogen-associated molecular patterns, or simply PAMPs [9]. Generic examples of PAMPs include lipopolysaccharides (LPS), lipoproteins on bacterial/fungal cell walls, single or double stranded RNA molecules, and most importantly unmethylated CpG motif containing DNA/synthetic oligodeoxynucleotides [7, 10]. Another group of components that are of not pathogenic but host origin is also recognized by PPRs; they are termed as damage-associated molecular patterns, or simply DAMPs. DAMPs are products of damaged cells, tissues or remnants of necrotic/apoptotic cells and act like immunostimulatory agents that activate immune system for the purpose of cleansing within the context of tissue repair [14].
1.4 Toll-Like Receptors (TLRs)
Cells of innate immunity recognize bacterial and viral elements by PPRs via PAMPs and differentiate between self and non-self [1, 2]. To date, the most extensively studied PPRs are TLRs due to the critical missions they have been undertaking as a part of their innately given list of tasks mentioned below [15].
Basically, TLRs are type I transmembrane proteins and the members of Interleukin-1 receptor (IL-1R) superfamily. Their discovery was made in fruit fly (Drosophila melanogaster) in early 1990s and named after the Drosophila gene, Toll (meaning super or fantastic), whose original function was identified a decade back in a totally irrelevant context. Initially, Toll receptor was thought to regulate embryonic development of the flies. A decade later, it was realized that this receptor is responsible for major antifungal immune response [16]. Its homologous receptors were identified as a part of
4
mammalian innate immune system by Medzhitov and Janeway in 1997 and referred to as Toll-like receptors [17].
TLR as a transmembrane protein contains two major structural sections; an ectodomain of leucine-rich repeats (LRRs) and Toll-IL-1 receptor, or simply TIR, domains. While LRRs enable PAMP recognition by means of the docking sites protruding from cellular or endosomal membrane, TIR domain maintains the downstream signaling pathway inside the cell. 12 different TLRs (TLR1-9 and TLR11-13) in mice and 10 different TLRs (TLR1-10) in human have been identified so far. Some of them (TLR1, 2, 4-6, 11, 12) are located at cell surface embedded in the membrane protruding outward to detect components like LPS before pathogenic invasion while some others (TLR3, 7/8, 9) reside inside the cell embedded in the membrane of intracellular vesicles such as endosomes, lysosomes, and endoplasmic reticulum (ER) to detect microbial DNA and/or RNA after invasion [3, 11, 13]. Table 1.1 shows the chromosomal locations of the genes that code for TLRs [3, 11].
Table 1.1. Chromosomal localizations of TLRs [3, 11].
TLR Mouse Human Chromosome TLR1 5 37.0 cM 4p14 TLR2 3 E3 4q32 TLR3 8 B2 4q35 TLR4 4 33.0 cM 9q32-q33 TLR5 1 98.0 cM 1q41-q42 TLR6 5 37.0 cM 4 4p14 TLR7 X F5 Xp22.3 TLR8 X F5 Xp22 TLR9 9 F1 3p21.3 TLR10 N/A 4p14 TLR11 14 C1 N/A TLR12 4 D2.2 N/A TLR13 X D N/A
TLRs play a pivotal role in; detecting danger signals proactively before things go bad, double checking to see whether the host is pathogen-free, directly activating innate immunity before adaptive immunity slowly takes over the “business” (which is a late-coming response in most of the occasions). TLRs have the potential to recognize a wide range of pathogens from viruses and bacteria through parasites and fungi (Table 1.2) [10, 13]. This versatility is generally attributed to the number of different types of TLRs present in host cell. Yet, it was also speculated that in TLR null mice
5
adaptive immunity could hold on its own without letting the host suffer irreversibly from infections [18].
Table 1.2. TLRs and their agonists [10, 13]. Types Cellular
localization Pathogens Agonists
TLR1/2 Membrane Bacteria Lipoprotein, LTA, PGN TLR2 Membrane Virus, Fungi, Parasites Structural Protein, Mannan,
Mutin TLR3 Endosome dsRNA, ssRNA, dsDNA,
Viruses dsRNA, poly(I:C), polyU TLR4 Membrane Bacteria, Viruses, Fungi,
Parasites
LPS, Structural Protein, Mannan,
Glycoinositolphospholipids
TLR5 Membrane Bacteria Flagellin
TLR2/6 Membrane Bacteria, Fungi Lipoprotein, LTA, PGN, Zymosan, B-glucan
TLR7/8 Endosome ssRNA Viruses, Bacteria, Fungi, Protozoan Parasites
GU-rich ssRNA, Imidazoquinolines (R848,
imiquimod, 3M001/2), Guanosine Analogues TLR9 Endosome ds DNA viruses, Bacteria,
Protozoan Parasites DNA, CpG ODNs, Hemozoin
1.4.1 Cell Surface Toll-Like Receptors 1.4.1.1 TLR1, TLR2, and TLR6
One of the most interesting TLRs is TLR2, which forms heterodimers with TLR1, TLR6, or with non-TLR molecules like dectin-1, CD14, and CD36 (Figure 1.1). This promiscuous interaction pattern provides TLR2 with a wide range of ligand recognition capabilities from bacteria and fungi through parasites and various species of viruses. TLR2-TLR1 and TLR2-TLR6 heterodimers recognize triacyl-lipopeptide (a membrane component of Gram (-) bacteria) and diacly-triacyl-lipopeptide (an essential element in mycoplasma membrane), respectively [10, 19, 20]. TLR2, when associated with dectin-1 (a C-type leptin receptor), recognizes zymogen. TLR2-TLR6 heterodimer senses LTA and MALP-2 (S.
aureus components) if and only if the co-receptor CD36 (class II scavenger receptor) get involved in
6 1.4.1.2 TLR4
TLR4 specialized to recognize lipopolysaccharide (LPS), the outer membrane component of Gram (-) bacteria that causes septic shock in human due to its extreme immunostimulatory potency [22]. LPS-TLR4 interaction depends heavily on the involvement of MD2 (a recognition subunit; myeloid differentiation protein-2) in complex formation with TLR4, LBP (LPS binding protein) and CD14 that is anchored in membrane bound GPI (glycosylphosphatidylinositol) [4]. In addition to LPS, TLR4 recognizes glycoinositolphospholipids of Trypanosoma [23] and fusion protein of RSV (respiratory syncytial virus), as well [24, 25].
1.4.1.3 TLR5
TLR5 recognizes a bacteria-specific structural component called flagellin, which makes up whip like appendage named as the flagella for bacterial movement [26]. Acting on TLR5 of epithelial cells and macrophages, flagellin triggers mucosal immune response. Highly conserved primary structure of flagellin makes it a great target for innate immunity. TLR5 is expressed ubiquitously; mainly in the respiratory tract, urinary tract, and gastrointestinal tract where inner wall epithelial cells are prone to bacterial infection any time. TLR5 activation results in chemokine expression from epithelial cells, which in turn adds to inflammatory responses and promotes leukocyte accumulation at the site of infection [27].
1.4.1.4 TLR11
TLR11 recognizes profiling-like molecules of T. gondii, an intracellular parasite of protozoan origin [28]. These molecules play a pivotal role in parasite motility and invasion [29]. Sensing the invader and establishing immune response against it within the cell is the first step to rid of this parasite, as one can imagine. Together with that, TLR11 provides an extra layer of protection against uropathogenic bacteria that might even cause kidney failures at later stages of infection if left unattended [30]. Interestingly enough, TLR11 is mouse-specific, not expressed in human due to the presence of a premature stop codon in the corresponding gene (Table 1.1) [11]. Whether or not or why this mutation is being naturally selected in human by evolution is a matter of discussion.
7
Figure 1.1. PAMP recognition by cell-surface TLRs (Adapted from [3])
1.4.2 Intracellular Toll-Like Receptors
As mentioned before, phagocytosis is a key process within the context of immunity where specialized cells of immune system directly engulf and degrade pathogens into monomers after a sequential order of events through endosomal pathway. After internalization and digestion of pathogens in the late endosome by the host, intracellular TLRs start interacting with nucleic acid motifs as a direct result of microbial cargo leakage into endosomal vesicles [18]. A wide range of microbes including viruses are sensed using non-self nucleic acid-specific TLRs inside the cell. Intracellular TLRs act like last line of defense before infection stabilizes once cell surface TLRs and other immune system mechanisms are breached by pathogens. That is exactly why these intracellular TLRs have been in the spotlight for a long time in the field. While TLR3 and TLR7/8 recognize single- and double-stranded RNA, TLR9 detects DNA of bacterial origin or single-stranded synthetic oligodeoxynucleotides (ODN) which are expressing unmethylated CpG motifs (CpG ODN) [3, 31-33]. Table 1.3 explains cellular localization of endosomal TLRs with a special emphasis on mouse and human orthologs.
8
Table 1.3. Cellular localization of endosomal TLRs [50 ,51].
Mouse Human TLR3 TLR7 TLR8 TLR9 TLR3 TLR7 TLR8 TLR9 pDC + + + + XCR1- DCs + + + XCR1+ DCs + + Monocytes + + + + + B cells + + + + + Neutrophils + + 1.4.2.1 TLR3
TLR3 recognizes double stranded RNA (dsRNA) and its synthetic analog polyinosinic-polycytidylic acid (poly(I:C)), which is used to mimic viral infection [31]. While TLR3 has been specialized to recognize RNA viruses such as reovirus in its natural setting, they also sense viruses with single stranded RNA (ssRNA) genome like Tobacco Mosaic virus and double stranded DNA (dsDNA) viruses like herpes simplex virus through expression of dsRNA that emerge during cellular transcription [34]. TLR3 is localized at the surface of early endosome embedded in the membrane as the drop of pH is vital for receptor-ligand engagement. TLR3 is unique in that, unlike TLR7/8 and TLR9, endoplasmic reticulum is not required for its intracellular trafficking [18].
1.4.2.2 TLR7 and TLR8
Both TLR7 and TLR8 recognize guanine analogs, ssRNA and its synthetic analogs imiquimods / resiquimods [32, 35, 36]. Some ssRNA viruses such as HIV and human influenza virus, along with certain bacteria species release their RNA content into intracellular endosomes after internalization and it is these RNA molecules that are the target for TLR7 and TLR8 [37, 38]. Interestingly enough, some siRNAs were also shown to get recognized by TLR7 [39]. Imiquimod acts as a TLR7 agonist that is now used as an FDA approved drug to treat neoplasias, lentigo maligna, and basal cell carcinoma due to its stimulation of pro-inflammatory cytokine and chemoattractant secretion and of Th1-based immune response [40]. Resiquimod (R848) is a potent agent that activates TLR7 and TLR8 together, yet causing excessive cytokine release in human as a part of its side effects in clinical use [41].
9 1.4.2.3 TLR9
By far the most extensively studied TLR is TLR9, which recognizes bacterial and viral genomic DNA by their unmethylated CpG repeats (those repeats are heavily methylated in mammals) [42, 43]. The immunostimulatory effects of these unmethylated CpG oligodeoxynucleotides (CpG ODNs) have been elucidated during studies that investigate ODNs on lymphocyte proliferation [44]. When CpG repeats were methylated or transformed into GpC, no immunostimulatory effect was observed in multiple experimental settings [42, 43]. Nevertheless, the mechanism through which TLR9 recognizes CpG ODN was uncovered some time later [45]. According to this explanation, TLR9 priming improves innate immune response along with Th1-based adaptive response via MyD88 (a key adaptor protein) recruitment and NF-kB/IRFs activation [46]. CpG ODN has a wide range of effects on immune system overall, including dendritic cell (DC) maturation, improved antigen presentation, increased production of pro-inflammatory cytokines, and elevated adaptive and humoral immune response when administered together with antigen.
Although TLR9 was engineered to distinguish between self and non-self DNA based on the methylation signature on CpG motifs in a sequence dependent manner, it might sometimes recognize self DNA as non-self and provoke immune system. When methylated DNA is mixed with LL37 or N-[1-(2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate (DOTAP) and immune cells are treated with that, the cells take up the “package” by endosomal pathway. Under these circumstances, endosomal DNA was shown to prime TLR9 [47-48]. To gain deeper insight into TLR9-CpG ODN interaction, recently elucidated 3D structure of this transmembrane protein with and without CpG ODN in its binding pocket can be examined [49]. The crystal structure showed that only CpG ODN-bound TLR9 could form a symmetric TLR9-CpG complex with 2:2 stoichiometry. It is this dimer complex that activates host immune system and ensures the continuity of the response till the danger is avoided. Within the scope of the same study, an inhibitory DNA molecule (iDNA) as TLR9 antagonist was also tested to see as to whether any ligand of DNA origin can cause TLR9 dimerization, and in turn, prime immune system. It was concluded that neither dimer complex formation was observed nor immune stimulation was noticed in the absence of CpG ODN. In this way, the mystery lying behind the structural basis of TLR9-CpG ODN interaction was uncovered with sufficient scientific proof.
10 1.4.2.4 Trafficking of Intracellular TLRs
As mentioned before, the localization of the intracellular TLRs (TLR3, TLR7, TLR8, and TLR9) are constrained to membrane-bound organelles such as endosomes, lysosomes, and ER [3, 13]. The importance of such specialized localization inside the cell can be emphasized in two different contexts. Firstly, viral mode of entry involves receptor-mediated endocytosis and their viral genomes are almost exclusively encapsulated within endosomes, which must have an early warning system to convey the bad news to the host. Secondly, intracellular TLRs expressed on the cell surface cannot differentiate between self and non-self DNA; it was shown that macrophages which express TLR9 on the cell membrane is responsive to DNA of self origin (implying induced autoimmunity) [11, 52]. These intracellular TLRs are originally present on ER membrane in the absence of stimulation and after PAMP insult they traffic immediately to endosomal compartments for proper activation (due to the structural need of intracellular TLRs for low pH environment) [11, 53].
Trafficking of intracellular TLRs is controlled by a membrane spanning protein on ER called UNC93B1. UNC93B1 cooperates with transmembrane domains of these TLRs to facilitate their re-localization from ER to the endolysosomes [54, 55]. It was revealed that “crippled” UNC93B1 leads to defective cytokine production and malfunctioning of co-stimulatory molecules involved when the cells are insulted using intracellular TLR ligands [56]. In fact, UNC93B1 is so critical for human immune system that its deficiency makes the host prone to HSV-1 encephalitis [57]. Figure 1.2 shows PAMP recognition by intracellular TLRs and the role of UNC93B1 in their trafficking from ER to endolysosome.
There are two other ER proteins that involve TLR trafficking, namely PRAT4 and GP96. However, they are not specific to intracellular TLRs; all TLR trafficking gets spatially affected from their absence. PRAT4A plays a central role in TLR1, TLR2, TLR4, TLR7, and TLR9 trafficking from ER to cell membrane or endolysosome while GP96 acts as a universal chaperone for proper folding of TLR1, TLR2, TLR4, TLR5, TLR7, and TLR9 during their journey inside the cell [58, 59]. As seen above, trafficking of both intracellular and cell surface TLRs involves the contribution of multiple elements for proper TLR localization, and in turn, their correct functioning. If a single player is taken out of the game, the whole system collapses, resulting in a mess of malfunctioning parts. So, it is vital to keep a healthy immune system that works for the host, not against it.
11
Figure 1.2. PAMP recognition by intracellular TLRs and the role of UNC93B1 in their trafficking from ER to endolysosome (Adapted from: [3])
1.4.2.5 Endosomal Cleavage of Intracellular TLRs
Acidification, mentioned above as low pH environment, is a must for intracellular TLR activation due to smooth functionality of these receptors. Two major reasons are behind this phenomenon. Firstly, acidic environment activates enzymes that facilitate dismantling of microbes in endolysosomes so that their content like unmethylated CpG DNA becomes physically accessible for immune activation [60, 61]. Secondly, processing to functionally activate of inactively produced TLRs involves protease cleavage, which occurs only at a low pH as in the case of many proteolytic cleavage reactions in cell. It is only the properly cleaved TLR9s that can recognize ligand and initiate signaling cascade downstream. It was suggested that some other TLRs like TLR7 also require pre-processing to become functionally active but not all (e.g. TLR3) [62], indicating that cleaved versions of TLR9 and TLR7 might reveal same binding affinity to their ligands as full-length TLR3 does to its own [18]. Overall, proteolytic pre-cleavage in TLR maturation process appears to be an extra layer of regulation towards proper functionality of the TLR-based immunity.
12 1.5 TLR Signaling
After an induced fit between PAMP and corresponding TLR, TLRs activate customized immune responses. To illustrate, some of the cell surface TLRs (namely TLR1-TLR2, TLR2-TLR6, and TLR5) promote production and secretion of inflammatory cytokines while TLR3 and TLR4 lead to both type I IFNs and inflammatory cytokine responses. This level of specificity, meaning the customized TLR responses to different pathogens, relies heavily upon proliferation and/or recruitment of a single or specific combination of the Toll/Interleukin-1 receptor (TIR) domain containing adaptor proteins, for instance Myeloid differentiation primary response gene 88 (MyD88), TIR domain containing adaptor protein (TIRAP), TIR-domain-containing adapter-inducing interferon-β (TRIF), and TRIF related adaptor molecule (TRAM) [63]. TLR1, TLR2, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9 and TLR11 call for MyD88 to their TIR domains once proper ligand-receptor interaction occurs at the docking site of the corresponding TLR. There exists a different path for TLR3 and TLR4. They recruit TRIF, in place of MyD88. TLR1, TLR2, TLR4 and TLR6 exploit TIRAP, which acts as bridge between their TIR domains and MyD88 while TLR5 and TLR11 interact directly with MyD88 without the need for another molecule. Likewise, TLR4 needs TRAM for a successful TRIF interaction. These special requirements of specific TLRs for different adaptor molecules converge into a few major signaling cascades, ultimately activating transcription factors like nuclear factor kappa B (NF-kB) and interferon regulatory factor (IRFs) [15] (Figure 1.3).
13
TLR signaling can be split in two; MyD88-dependent pathway and TRIF-dependent pathway. The only TLR that can possibly use all four existing adaptor molecules mentioned above and that is capable of inducing both pathways is TLR4 [3].
1.5.1 MyD88-dependent Pathway
MyD88-dependent pathway is a generic one; all known TLRs with the exception of TLR3 exploit it. After an induced fit between TLR and its ligand, TLRs immediately call for MyD88 molecules to their TIR domains. A sequential order of events after that ensures successful completion of this signaling cascade. MyD88 recruits elements of IL1 receptor associated kinase family, and in turn, TNF receptor associated factor (TRAF) 6. TRAF6 promotes TAK1, which in turn activates; 1. IκB kinase (IKK) complex to subsequently activate NF-kB and, 2. mitogen-activated protein kinase (MAPK) pathway [15] (Figure 1.4).
14 1.5.2 TRIF-dependent Pathway
This signaling pathway is used only by TLR3 and TLR4 after excitation by ligand binding. This particular pathway leads to production and secretion of type I IFNs and inflammatory cytokines in DCs and macrophages. TRIF recruits TBK1, the kinase protein that phosphorylates IRF3, with the help of TRAF3. Once phosphorylated, IRF3 travels from cytoplasm to nucleus and leads to production of type I IFNs. In plasmacytoid dendritic cells (pDC) a shortcut of this pathway exists; MyD88 involves direct interaction with IRF7 to induce immediate type I IFN production [27] (Figure 1.4). Considering that pDCs circulate in blood and function primarily in peripheral lymphoid organs, this time-saving shortcut makes biological sense; newly identified pathogens that managed to penetrate through physical barriers of the host should be dealt with in no time before it is too late to take action against them.
1.6 Effects of DNA on Immune System
Together with the conventional role DNA and RNA play in information storage and inheritance as blueprints of life, they have an additional and almost totally irrelevant task to undertake; these nucleic acids as physical entity modulate immune system [64-66]. After microbial infection, genomic content of the invader or the damaged host cells that is perfectly sealed within bilayer or coat proteins is released outside and expose to all kinds of foreign particles ranging from lipid vesicles and intracellular proteins through adaptor molecules and extracellular receptors. This genomic content is detected by the immune system [24]. On one side, DNA of bacterial origin, for instance, are recognized as non-self by host through TLR9 receptors because of its high level of unmethylated CpG content and this recognition initiates an innate immune response defined as the expansion and maturation of plasmacytoid dendritic cells (pDC), natural killer cells, and ultimately B lymphocytes along with the release of Th1-type extracellular proteins such as cytokines, chemokines, and/or multivalent immunoglobulins [46, 67-69].
On the other side, DNA of mammalian origin has a totally opposing effect on immune system when released out of the nucleus. The tips of mammalian chromosomes called telomeres contain immunosuppressive TTAGGG repeats that can curb immune response. This response is generally Th1-mediated but not always TLR-dependent. Interestingly enough, TTAGGG repeats in non-eukaryotic genome seem to be highly underrepresented, implying the evolutionary advantage provided by an additional mechanism of immune evasion for pathogens [22, 69-71]. As one can imagine, the implications of these immunosuppressive telomeric motifs in clinics within the context of autoimmunity are indeed beyond measure. Together with immunostimulatory CpG DNA, they
15
were examined below in detail both theoretically and practically in the mainstream of this research study.
On a separate not, novel therapeutics that mimic immunomodulatory effects of both immunostimulatory and immunosuppressive DNA motifs are already under the (pre)clinical trials to treat a wide range of diseases from cancer through autoimmunity [72-74]. More specifically, the synergistic interaction between unmethylated CpG DNA and TLR9 has been successfully mimicked using synthetically-made oligodeoxynucleotides (ODN) bearing bacterial CpG DNA motifs with the ultimate goal to include it within vaccine formulations as immune adjuvant and to yield more effective and prolonged immunity against disease in human [75-77]. Additionally, there exists artificially-produced immunoinhibitory motifs of telomeric origin which can suppress immune system to fine-tune delicate balance of immune response against pathogens; an overly active immune response is just as harmful to the host itself, as in the case of autoimmune disorders [78]. Throughout the period from initial mouse studies where primary findings were implemented into practice first time ever through the current human clinical trials the use of immunomodulatory ODNs in clinics has gained popularity increasingly.
1.6.1 Immunostimulatory CpG ODN
The initial observation that bacterial genome has anti-tumor effects has led to a deep down investigation of what might have caused this peculiarity [67]. It was understood later on that the sequences of unmethylated CpG flanked by two purines and two pyrimidines on the 5’ and 3’ ends, respectively initiate a major immune response where Th1-mediated cytokines and pro-inflammatory chemokines are all involved [44]. It became clear after some years the content, the length, the methylation state, and the number of CpG motifs present in a molecule of CpG ODN have direct impact on CpG ODN’s ability to prime host immunity [79]. For instance, those ODNs which harbor unmethylated Cytosine-Guanine nucleobases within a motif of Purine-Purine-CpG-Pyrimidine-Pyrimidine from 5’ end through 3’ were found to be rather successful in activating the immune system. The level of success achieved is directly proportional to the number of CpG nucleobase pairs present in ODN; the more the number of CpG dinucleotides added, the higher the immune response observed [75].
The presence of at least one unmethylated CpG motif is a hallmark of CpG ODNs. It is their flanking regions that enable us to distinguish between two major classes; D-type (also called A-class) and K-type (also known as B-class). A newly-identified class called C-class CpG ODNs is also of importance,
16
as described below. D-type ODNs are known to accommodate poly G-tail that let them form nanoparticle-like structures while K-type ODNs are more than often characterized by the presence of two or more CpG motifs [80]. D-type ODNs retain their integrity after internalization due to their rigid structure kept even inside the early endosome where it interacts with MyD88/IRF7 complexes and leads to INFα secretion by pDCs. This is indeed a hallmark of D-type ODN induced immune response [81]. Different from D-type ODN, K-type CpG ODNs induces an immune response that is characterized by pDC origin TNFα secretion at large quantities due mainly to late endosome relocation inside the cell [82]. Figure 1.5 outlines immunostimulatory pathways followed by these two CpG ODNs inside the cell. Finally, the newly-identified class C-class ODNs has increasingly gained popularity in the field on the grounds that it combines the advantages provided by two main classes even though this synergy occurs at a lower level [83].
Figure 1.5. D-type (CpG-A) and K-type (CpG-B) ODN induced signaling pathways and their intracellular localization (Adapted from [82])
1.6.2 Immunosuppressive ODN
Inflammation caused by host’s own immune cells during infection or tissue wounds has detrimental effects on the surrounding tissues even though it has originally emerged to improve host’s odds of
17
ridding pathogens or damaged cells and still works perfectly for this purpose in a healthy system. So, inflammation must be eventually terminated with tissue remodeling during recovery. Similarly, other immune mechanisms originally designed to protect the host might lead to serious problems unless properly handled as in the case of autoimmunity [84]. It was recently found that in case of a possible cell burst after infection host DNA harbors immunosuppressive elements by entity capable of downregulating immune response against pathogen derived CpG rich DNA in an attempt to alleviate DNA-driven immune stimulation [85]. Similarly, during tissue damage the release of these immunosuppressive elements as host cell die may serve to diminish pathologic inflammatory and autoimmune response [86]. It is now known that neutralizing or suppressive ODNs can even put a halt on CpG-mediated immune stimulation in a selective manner [87]. It turned out that optimal sequence of an immunosuppressive motif is identical with telomere sequences (that seal the tips of mammalian chromosomes as a physical protection against exonuclease digestion or simply shortening), i.e. repeats of “TTAGGG” [64]. Gursel and his colleagues revealed for the first time that telomeric repeats are capable of inhibiting immune activation effectively. Other mammalian sequences that are non-telomeric failed in the attempt. They also obtained the same result in a telomerase defective KO mouse model, which could not suppress immune response in face of naturally-occurring or induced activation of immunity [64]. That was indeed a breakthrough which added a new line to our understanding of list of tasks telomeres originally undertake.
A follow-up research showed in vitro that telomeric suppressive ODN of mammalian origin, named A151, inhibits the production of many pro-inflammatory molecules induced by bacteria like IL6, IL12, IFNγ, TNFα and MIP2α [64, 88]. Table 1.4 and Table 1.5 quantify the effect of suppressive ODN on cytokine/chemokine production and induced expansion of immune cells [88].
Table 1.4. The effects of suppressive ODN on CpG-induced immune activation (Adapted from [88]).
Treatment % Activation CpG ODN alone 100±2 +Mammalian DNA 27±4 +Telomeric DNA 13±3 +Non-telomeric DNA 87±5 +Control ODN 97±3
+Suppressive ODN (A151) 8±2
18
Table 1.5. The effect of Suppressive ODN on IFNγ production induced by several immune activators (Adapted from [88]).
Stimulus No Treatment Control ODN Suppressive ODN (A151) CpG DNA 27±5 24±6 6±2 ds RNA 8±2 10±1 2±1 Peptidoglycan 28±5 31±4 5±2 LPS 24±7 21±3 6±2
In search for possible use of suppressive ODN (A151) in the treatment of autoimmune and inflammatory diseases, Gursel and his colleagues demonstrated that suppressive ODN administration to BALB/c mice right before LPS challenge substantially ameliorated survival rate statistics [22]. In another study suppressive ODN (A151) treatment significantly delayed the onset of kidney inflammation in NBZ/W mice with greatly prolonged survival rates [89]. Yet another study showed that A151 treatment considerably diminished the severity of arthritis in lab mice by reducing the serum titer of pathogenic IgG autoantibodies and IFNγ production by the corresponding T cells [90]. Furthermore, A151 was shown to alleviate the symptoms of pulmonary inflammation in BALB/c mice significantly [91]. Fujimoto et al. reported that ocular inflammation can be avoided in murine models when A151 is used to treat the inflammation [92]. 2011 was a remarkable year in terms of suppressive ODN research. Klinman and his colleagues used A151 in the treatment of a particular type of cancer characterized by major inflammations across the host body [93]. The outcome was strikingly positive; A151 blocked hyperplasia, edema, and leukocyte infiltration which are vital to tumorigenesis. This was the first attempt to use suppressive ODNs in inflammation-associated oncogenesis treatment, which yielded optimistic results for future research. In a follow-up study, Klinman and again his colleagues reported that A151 is capable of reducing lung cancer susceptibility of mice with inflammatory disorders [94]. Overall, the A151 research is still on the spotlight and seems to continue to do so for a long time.
Structurally speaking, A151 is a telomeric sequence composed of poly G runs. These guanine runs, by definition, are prone to establish inter- and intra-chain Hoogstein bonds in their thermodynamically most favorable forms called G-tetrads. Gursel and his colleagues demonstrated that suppressive properties of A151 can be at least partially attributed to its ability to form these G-tetrads [64]. It was shown in the same work that any nucleotide interfering with G-tetrad formation can cease A151’s ability to suppress immunity.
19
Suppressive ODN A151 has diverse effect on components of immunity [86] (Figure 1.6). It supports proliferation of Th17 and iTregulatory cells with immunosuppressive activity. It suppresses activated Th1 cells, resulting in a Th2 bias in the following responses. A151 hampers continued activation of dendritic cells and macrophages, leading to diminished production of pro-inflammatory molecules. Other A151 effects include suppression of B cell maturation, class switching, and Ig production. So, it appears that A151 holistically suppresses immune response using a wide range of interdependent mechanisms.
Figure 1.6. Diverse effects of suppressive ODN A151 on cellular elements of the immune system
(Adapted from [86])
Even though A151 is by far the most promising and most effective immunosuppressive ODN identified so far for future application, it is not the only ODN that has such effects on immunity. Most commonly used ODNs after A151 with similar effects on immune system are H154 (CCTCAAGCTTGAGGGG) [69], INH ODN (TCCTGGCGGGGAAGT) [95], modified CpG ODN
(TGACTGTGAAGGTTAGAGATGA, TCCATGAGCTTCCTGATGCT) [96], S ODN
(GGGGGGGGGGGGGGGGGGGG) [97], IRS ODN (TGCTCCTGGAGGGGTTGT) [98], G ODN (CTCCTATTGGGGGTTTCCTAT) [95], and microsatellite ODN (AAAGAAAGAAAGAAAGAAAGAAAG, CCTCCTCCTCCTCCTCCTCCTCCT) [71]. Each of these sequences has different merits and caveats for medical use but, compared to A151, their immunosuppressive potencies within different contexts that are immune relevant appear to be questionable. An important insight based on their sequence content, though, is that majority of these sequences have poly G runs and that the
![Table 1.3. Cellular localization of endosomal TLRs [50 ,51].](https://thumb-eu.123doks.com/thumbv2/9libnet/5689006.114876/27.892.120.783.143.358/table-cellular-localization-of-endosomal-tlrs.webp)
![Table 1.4. The effects of suppressive ODN on CpG-induced immune activation (Adapted from [88])](https://thumb-eu.123doks.com/thumbv2/9libnet/5689006.114876/36.892.225.678.880.1052/table-effects-suppressive-odn-induced-immune-activation-adapted.webp)
![Table 1.5. The effect of Suppressive ODN on IFNγ production induced by several immune activators (Adapted from [88])](https://thumb-eu.123doks.com/thumbv2/9libnet/5689006.114876/37.892.230.663.173.319/table-effect-suppressive-ifnγ-production-induced-activators-adapted.webp)
![Figure 1.6. Diverse effects of suppressive ODN A151 on cellular elements of the immune system (Adapted from [86])](https://thumb-eu.123doks.com/thumbv2/9libnet/5689006.114876/38.892.155.752.330.728/figure-diverse-effects-suppressive-cellular-elements-immune-adapted.webp)
![Figure 1.7. Major pathways of replicative and stress-induced premature senescence (Adapted from [122])](https://thumb-eu.123doks.com/thumbv2/9libnet/5689006.114876/44.892.231.667.99.618/figure-major-pathways-replicative-induced-premature-senescence-adapted.webp)