i
SLOW RELEASE AND DELIVERY OF ANTISENSE OLIGONUCLEOTIDE DRUG BY SELF-ASSEMBLED PEPTIDE
AMPHIPHILE NANOFIBERS
A THESIS
SUBMITTED TO THE MATERIALS SCIENCE AND NANOTECHNOLOGY
PROGRAM OF GRADUATE SCHOOL OF ENGINEERING AND SCIENCE OF BILKENT UNIVERSITY
IN PARTIAL FULFILLMENT OF THE REQUIREMENTS
FOR THE DEGREE OF
MASTER OF SCIENCE
By
SELMA BULUT
ii
I certify that I have read this thesis and that in my opinion it is fully adequate, in scope and in quality, as a thesis of the degree of Master of Science.
………
Assist. Prof. Dr. Ayşe Begüm Tekinay (Advisor)
I certify that I have read this thesis and that in my opinion it is fully adequate, in scope and in quality, as a thesis of the degree of Master of Science.
……….
Dr. Turgay Tekinay, Assist. Prof. Dr. Mustafa Özgür Güler (Co-Advisor)
I certify that I have read this thesis and that in my opinion it is fully adequate, in scope and in quality, as a thesis of the degree of Master of Science.
………. Prof. Dr. Engin U. Akkaya
I certify that I have read this thesis and that in my opinion it is fully adequate, in scope and in quality, as a thesis of the degree of Master of Science.
………. Assist. Prof. Dr. Fatih Büyükserin
Approved for the graduate school of engineering and science: ……….
Prof. Dr. Levent Onural
iii
ABSTRACT
SLOW RELEASE AND DELIVERY OF ANTISENSE OLIGONUCLEOTIDE DRUG BY SELF-ASSEMBLED PEPTIDE
AMPHIPHILE NANOFIBERS
Selma Bulut
M.S. in Materials Science and Nanotechnology
January, 2012
Antisense oligonucleotides are short single stranded DNA sequences and they are suggested to be used for treatment of several disorders including cancer. They could enter the cell and specifically inhibit the target gene, however chemical stability, controlled release and intracellular delivery are areas that has to be focused on to increase their efficacy. Gels composed of nanofibrous peptide network have been previously suggested as carriers for controlled delivery of drugs to improve stability and to provide controlled release, but have not been used for oligonucleotide delivery. In this work, a self-assembled peptide nanofibrous system is formed by mixing a cationic peptide amphiphile (PA) with Bcl-2 antisense oligodeoxynucleotide (ODN), G3139, through electrostatic interactions. The self-assembly of PA-ODN gel was characterized by circular dichroism, rheology, atomic force microscopy (AFM) and scanning electron microscopy (SEM). AFM and SEM images revealed establishment of the nanofibrous PA-ODN network. Due to the
iv
electrostatic interactions between PA and ODN, ODN release can be controlled by changing PA and ODN concentrations in the PA-ODN gel. Cellular delivery of the ODN by PA-ODN nanofiber complex was observed by fluorescently labeled ODN molecule. Cells incubated with PA-ODN complex had enhanced cellular uptake compared to cells incubated with naked ODN. Furthermore, Bcl-2 mRNA amounts were lower in MCF-7 human breast cancer cells in the presence of PA-ODN complex compared to naked ODN and mismatch ODN evidenced by quantitative RT-PCR studies. These results suggest that PA molecules can control ODN release, enhance cellular uptake and present a novel efficient approach for gene therapy studies and oligonucleotide based drug delivery. In follow-up studies, increase in the internalization efficacy of ODN by incorporation of bioactive sequences, RGDS, to peptide sequence was also shown.
Keywords: peptide; amphiphile; nanofiber; antisense oligonucleotide; G3139; Bcl-2; drug delivery; anticancer drug
v ÖZET
DNA BAZLI ANTİKANSER İLAÇLARININ NANOFİBERLERLE KONTROLLÜ SALIMI VE TAŞINIMI
Selma Bulut
Malzeme Bilimi ve Nanoteknoloji Programı, Yüksek Lisans
Ocak, 2012
Antisens oligodeoksinükleotitler (ODN) kanser dâhil birçok hastalığın tedavisinde kullanılabilen önemli ilaçlardır. Ancak kimyasal dayanıklılık, kontrollü salım ve hücre içerisine alım oligonükleotitlerin verimini etkileyen önemli faktörlerdir. Nanofibrillerden oluşan üç boyutlu ağsı yapıya sahip jeller ilaçların kontrollü salımı ve taşımı amacıyla ilaç dayanıklılığını arttırmak için kullanılmıştır, fakat oligonükleotitler için bir çalışma yoktur. Bu çalışmada, programlı toplanabilir peptit nanofiber sistemi, pozitif yüklü amfifilik peptit (AP) molekülleri ile Bcl-2 antisens oligodeoksinükleotidi, G3139, karıştırılarak elektrostatik etkileşimlerin etkisiyle oluşturuldu. Programlı toplanabilir AP ve ODN jeli dairesel dikroism, reoloji, atomik kuvvet mikroskobu (AFM) ve taramalı elektron mikroskobu (SEM) kullanılarak karakterize edildi. AFM ve SEM görüntüleri jelin nanofiber ağsı yapısını göstermektedir. Jelden ODN
vi
salımı katyonik AP‟ler ve anyonik ODN‟ler arasındaki elektrostatik etkileşimlere bağlıdır. Çalışmamızda AP ve ODN konsantrasyonları değiştirilerek ODN salımı kontrol edildi. Floresan işaretli ODN kullanılarak AP-ODN kompleksinin hücre içerisine alınımı incelendi. AP-AP-ODN kompleksi uygulanan MCF-7 insan meme kanseri hücrelerinin daha fazla floresan sinyali verdiği gözlendi. Ayrıca AP-ODN kompleksi ile inkübe edilen MCF-7 hücrelerinin Bcl-2 mRNA ifadesinin, çıplak halde ODN ve 2-bazı eşleşmeyen (mismatch-MM) ODN, 2-bazı eşleşmeyen (mismatch-MM) ODN ve AP kompleksi ile inkübe edilen hücrelerdeki mRNA ifadesinden daha fazla azaldığı kantitatif RT-PZR yöntemi ile gösterildi. Bu sonuçlar AP moleküllerinin ODN salımının kontrolü ve hücre içerisine verimli ODN alımı için kullanılabileceğini ve gen terapisinde yeni bir yaklaşım olabileceğini önermektedir. Devam eden çalışmalarda, ODN„nin hücre içerisine alımının RGDS biyoaktif dizileriyle arttırıldığı gösterildi.
Anahtar Kelimeler: peptit; amfifil; nanofiber; antisens oligonukleotit; G3139; Bcl-2; ilaç taşınımı, antikanser ilaç
vii ACKNOWLEDGEMENT
I am heartily thankful to my supervisor, Dr. Turgay Tekinay, for his encouragement, guidance and support during the course of this research. I would like to express my special thanks to my co-advisors; Dr. Mustafa Ö. Güler and Dr. Ayşe Tekinay for their support and sharing their knowledge.
I would like to thank to Sıla Toksöz, Selman T. Erkal, and Yavuz Selim Dağdaş for their partnership in this research. I owe my deepest gratitude to Zeynep Ergül Ülger and Büşra Mammadow for their guidance and support from the initial to the final day of my study.
I want to thank to my group members; Turgay Çakmak, Burcu Gümüşçü, Diren Han, Pınar Angün, Özgün C. Onarman, Ömer Faruk Sarıoğlu and all members of Nanobiotechnology Lab; Ayşegül Tombuloğlu, Hakan Ceylan, Hilal Ünal, Reşad Mammadov, Samet Kocabey, Seher Üstün and Biomimetic Lab; Zeliha Soran, Ruslan Garifullin, Selim Sülek, Handan Acar, Oya Ustahüseyin and Rukan Genç. It was wonderful to work with them. Kızçeler; Aslı Çelebioğlu, Fatma Kayacı, Zeynep Aytaç, thank you for your unselfish and unfailing friendship.
I would like to thank to UNAM (National Nanotechnology Research Center) and TÜBİTAK (The Scientific and Technological Research Council of Turkey) grant number 110S018, for their financial support.
Finally, I want to express my gratitude to my family for their love, support, and understanding. I cannot thank you enough.
viii
Canım Ailem’e
ix LIST OF ABBREVIATIONS
PA: Peptide Amphiphile
ODN: Oligodeoxynucleotide
AS: Antisense
ECM: Extracellular Matrix
FMOC: 9-Fluorenylmethoxycarbonyl
FAM: Fluorescein amidite
BOC: ter. Butoxycarbonyl
MTT: 4-Methyltrityl
HBTU: 2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
DIEA: N, N-Diisopropylethylamine
DMF: Dimethylformamide
TFA: Trifluoroacetic Acid
LC-MS: Liquid Chromatography-Mass Spectrometry
AFM: Atomic Force Microscopy
SEM: Scanning Electron Microscopy
CD: Circular Dichroism
x DNA: Deoxyribonucleic Acid
RNA: Ribonucleic Acid
PNA: Peptide Nucleic Acid
xi TABLE OF CONTENTS ABSTRACT ACKNOWLEDGEMENTS TABLE OF CONTENTS LIST OF FIGURES 1. INTRODUCTION ... 1
1.1 Antisense Oligonucleotide Technology ... 1
1.2 Use of Peptide Amphiphile Nanofibers in Medicine ... 6
3.1 Peptide Amphiphiles for Release and Delivery of Antisense ODN……….10
EXPERIMENTAL SECTION ... 14
2.1. Materials ... 14
2.2. Antisense Oligonucleotides ... 14
2.3. Cells ... 15
2.4. Peptide Synthesis, Purification and Characterization ... 15
2.4.1. Synthesis of Peptide Amphiphiles ... 15
2.4.2. Characterization of Peptide Amphiphiles ... 16
xii
Viscoelasticity and Gelation Behavior ... 16
Morphological Observation ... 17
Atomic Force Microscopy (AFM) ... 17
Scanning Electron Microscopy (SEM) ... 18
2.5. ODN Release from PA-ODN Gel ... 18
2.6. Confocal Fluorescence Recovery after Photobleaching of FAM-Labeled ODN in PA-ODN nanofibrous network ... 19
2.7. In Vitro Studies ... 20
2.7.1. Cellular Uptake of PA-ODN Complexes ... 20
2.7.2. Cell Viability and Proliferation Assay ... 21
2.7.3. Determination of Bcl-2 mRNA Expression by Quantitative RT-PCR………..22
2.7.4. Western Analysis ... 24
RESULTS AND DISCUSSION ... 26
3.1. Peptide Synthesis, Purification and Characterization ... 26
3.2. ODN Release from PA-ODN Gel ... 37
3.3. In Vitro Studies ... 43
3.4. Future Perspectives ... 52
CONCLUSION ... 60
xiii LIST OF FIGURES
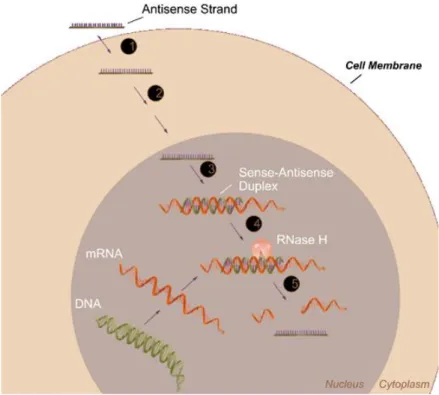
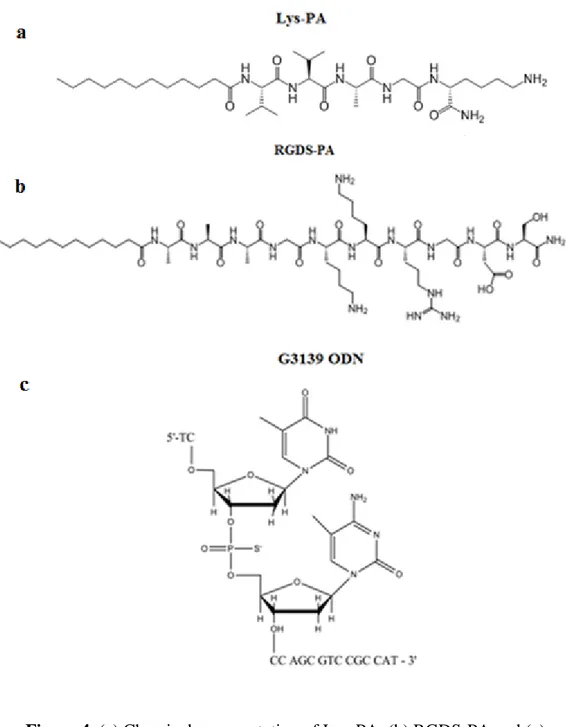
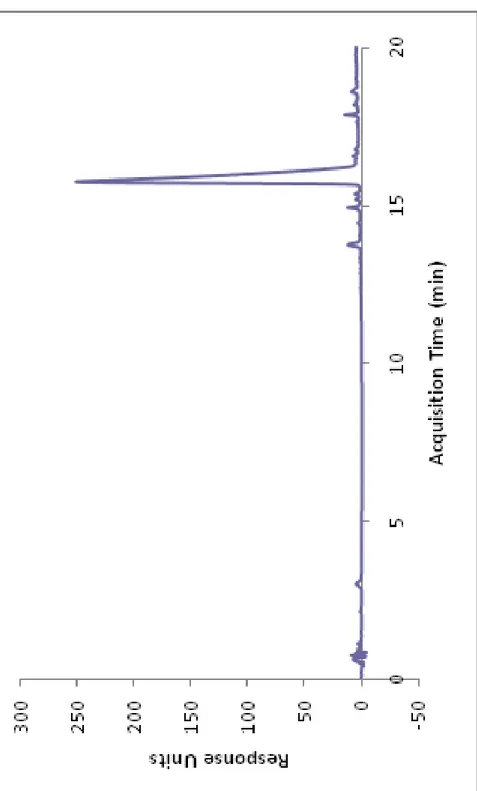
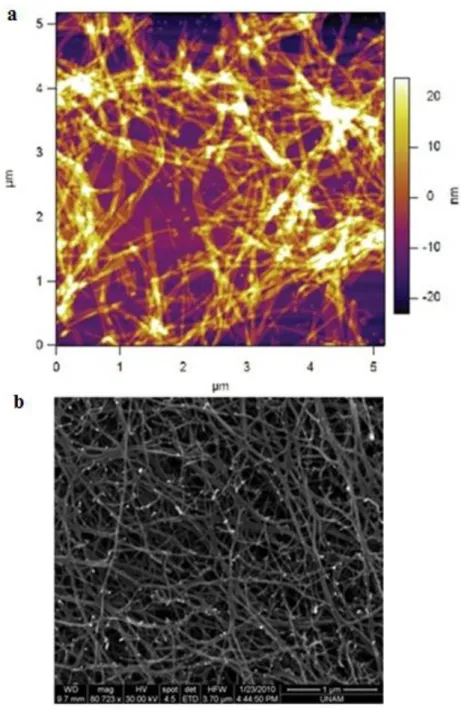
Figure 1. Representative image of RNase H-dependent antisense mechanism ... 4 Figure 2. Mechanism of action for Genasense®. Reproduced with permission from Genta Inc., NJ ... 13 Figure 3. Solid Phase Synthesis Diagram. Reproduced with permission from Sigma-Aldrich. ... 28 Figure 4. (a) Chemical representation of Lys-PA, (b) RGDS-PA and (c) Chemical structure of the backbone of the phosphorothioate G3139. ... 29 Figure 5. Mass spectrometry of the Lys-PA. ... 30 Figure 6. RP-HPLC chromatogram of the Lys-PA. ... 31 Figure 7. Schematic representation of the fibrous gel of Lys-PA (2 wt %) and Fluorescein tagged-ODN (1000 ng/µl) ... 32 Figure 8. CD spectra demonstrate increased β-sheet formation in PA- ODN mixture compared to PA alone ... 33 Figure 9. (a) AFM topography and (b) SEM images of nanofibrous 3-D network of the Lys-PA and ODN complex ... 34 Figure 10. Frequency sweep profiles of gels of Lys-PA (2 wt %) with ODN at different final concentrations (75-250 µM). ... 35
xiv
Figure 11. Storage modulus (G′) and loss modulus (G′′) of gels of Lys-PA (2 wt %) with varying concentrations of ODN at angular frequency of 100 rad/s. ... 36 Figure 12. Release profile shows of ODN from peptide nanofiber (a) gel of 30 µg/µl ODN and 2 wt % PA for 6 days, (b) gel of 0.2 µg/µl ODN and 2 wt % PA for 3 days. (c) Representative image of gel with PBS used for release experiments ... 40 Figure 13. Release profile of ODN from (a) gel of 0.1%, 0.15% and 0.2 wt% PA and ODN at concentrations of 150 ng/µl, (b) 300 ng/µl, (c) 600 ng/µl, and (d) 1200 ng/µl for 5 days. ... 41
Figure 14. Intensity graph of area of fluorescein ODN area at time of pre-bleach, post bleach and post-recovery compared to unbleached area ... 42 Figure 15. Representative images of MCF-7 cells treated with (a) naked FAM labeled ODN (green); (b) FAM labeled ODN and 0.01 wt% PA complex for 4 h followed by an additional incubation of 44 h without treatment at 37 oC. TO-PRO-3® (blue) was used to stain the nuclei. Images were taken at 63X magnification ... 47 Figure 16. The proposed mechanism of PA – ODN complex ... 48 Figure 17. (a) Viability and (b) proliferation assay results of MCF-7 cell line incubated with naked ODN/MM (mismatch) and complex with Lys-PA. ... 49 Figure 18. Change in gene expression of Bcl-2 in MCF-7 breast cancer cells incubated with PA-ODN (G3139)/mismatch (G4126) complexes and
xv
free ODN/mismatch. Total RNA was extracted from cells after 4 h of treatment followed by (a) 44 h (b) 68 h of incubation without treatment. GAPDH was used as housekeeping gene to normalize the gene expression level using Pfaffl method. ... 50 Figure 19. Western blot analysis of Bcl-2 protein expression in MCF-7 cells incubated for 48 h with Lys-PA - ODN complex ... 51 Figure 20. Western blot analysis of Bcl-2 protein expression in MCF-7 cells incubated for 72 h with Lys-PA - ODN complex ... 51 Figure 21. Mass spectrometry of the RGDS-PA ... 54 Figure 22. RP-HPLC chromatogram of the RGDS-PA. ... 55 Figure 23. Schematic representation of the RGDS-PA (0.04 wt %) and Fluorescein tagged-ODN (200 ng/µl) complexes ... 56 Figure 24. CD spectra of RGDS-PA and ODN mixture compared to PA alone at pH 6 and pH 10. ... 57 Figure 25. AFM topography images of the RGDS-PA and ODN complexes ... 58 Figure 26. Representative images of MCF-7 cells treated with (a) naked FAM labeled ODN (green); (b) FAM labeled ODN and 1 wt % RGDS-PA complex for 4 h followed by an additional incubation of 44 h without treatment at 37 oC. TO-PRO-3® (blue) was used to stain the nuclei. Images were taken at 63X magnification ... 59
1
Parts of this study was published as “Slow Release and Delivery of Antisense Oligonucleotide Drug by Self-Assembled Peptide Amphiphile Nanofibers” Selma Bulut , Turan S. Erkal , Sila Toksoz , Ayse B. Tekinay , Turgay Tekinay , and Mustafa O. Guler Biomacromolecules, June 28, 2011 (Web)”, Reproduced (or 'Reproduced in part') with permission from American Chemical Society. Copyright 2011 Royal Society of Chemistry.
1. INTRODUCTION
1.1 Antisense Oligonucleotide Technology
Antisense (AS) oligonucleotides are unmodified or chemically modified short, single stranded DNA or RNA sequences that are designed to bind specifically to target RNA1. Antisense oligonucleotides could be used as inhibitors to regulate expression of specific genes to downregulate target proteins for treatment of diseases such as cancer, viral infections and inflammatory diseases based on complementation of specific oligonucleotide to a chosen sequence2-3.
On the basis of the mechanism of action, there are two classes of antisense oligonucleotides, the steric-blocker oligonucleotides and the RNase H-dependent oligonucleotides. The steric-blocker oligonucleotides include methylphosphonates, 2′-O-methyloligoribonucleotides, PNAs
2
(Peptide Nucleic Acid), and morpholino oligonucleotides, which prevent protein expression by hybridizing with splice sites or intron regions of RNA physically.
The RNase H-dependent oligonucleotides have RNase H competent backbones that are phosphodiesters and phosphorothioates. Phosphodiesters and phosphorothioates are oligodeoxynucleotide (ODN) having sequences of 16 to 29 bases of single stranded DNAs. ODNs are the most common antisense oligonucleotides designed to specifically downregulate targeted genes by inhibiting translation of unwanted proteins by blocking the ribosomal machinery and/or recruiting endogenous RNase H enzyme6. After single-stranded ODNs enter the cell cytoplasm, they accumulate in the cell nucleus and hybridize with the complementary region in the corresponding RNA via Watson-Crick base pairing. DNA/RNA duplex triggers a defense mechanism and then RNase H, a ubiquitous enzyme, recognizes the oligonucleotide/RNA duplex as a substrate. RNase H cleaves the RNA strand and releases the antisense oligonucleotide that is free to bind to other targeted RNA molecules. RNase H is also present in the cytosol and cleaves DNA/RNA hybrids and down-regulates mRNA expression of specific proteins4-5(Fig. 1). Phosphorothioates, which contain sulfur in place of oxygen in the phosphodiester bond, are the most widely studied oligonucleotides. Because of their nuclease stability and relative ease of synthesis1, the phosphorothioate AS oligonucleotides are currently available for many clinical uses7,8,9-11. Fomivirsen (Vitravene) is a phosphorothioate
3
antisense oligonucleotide that has been approved by the Food and Drug Administration for treatment of cytomegalovirus retinitis3.
Due to ability to recognize specific target complementary sequences; in theory, antisense molecules can be designed to inhibit expression of any gene12. Use of oligonucleotides as gene expression inhibitors have some advantages, such as target specificity and reduced side effects. However, because of their rapid degradation by intracellular endonucleases and exonucleases, AS oligonucleotides cannot be used effectively as drugs to inhibit target gene expression13-14. To overcome these problems, there are ongoing studies to increase the stability of these molecules, such as chemical modification. The mostly used chemically modified oligonucleotides are methylphosphonates, 2′-O-methyloligoribonucleotides, PNAs, morpholino oligonucleotides and phosphorothioates6. Chemically modified oligonucleotides have improved stability, however, when they are not combined with proper delivery agents, they have limited ability to cross the cell membrane due to their charge and high molecular weight15,16. Thus, further developed and more efficient delivery systems are necessary for enhanced cellular uptake. To achieve sustained pharmacological activity of oligonucleotides, facilitate their delivery and to avoid multiple administrations, several release and non-viral delivery systems based on biodegradable polymers, lipids and their combination have been suggested such as cationic liposomes17-22, protamine-containing lipid nanoparticles23, synthetic biodegradable polymers, nano or microparticles (polylactide-co-glycolide microparticles)24-25, cationic nanoparticles26, hydrogels
(PLA-b-4
PEG-b-PLA hydrogel)27, micelles (polylactide–polyethylene glycol copolymers)28, fibrous scaffolds (electrospun polyethylene co-vinylacetate), polylactic acid29) and cationic polymers (polyethylenimine30, modified PEI/ODN polyplexes31, poly-lysine32, cationic facial amphiphiles33), and polycationic peptide34. These non-viral delivery systems are usually hydrophilic, positively charged and able to interact with oligonucleotides through electrostatic interactions permitting desired molecular design with controlled size for specific applications.35-36
Figure 1. Representative image of RNase H-dependent antisense mechanism (picture adapted with permission from Dean et. al.5).
5
In viral systems, human pathogenic viruses such as retrovirus, adenovirus, and baculovirus vectors are used as delivery agents with high transfection efficacies. However viral systems have limitations such as virally-induced inflammatory responses, immune reactions, finite capsule size with low performance in serum and costly process to maintain. As delivery agents, non-viral delivery vectors; lipids and/or cationic polymers and peptides, constructed with biodegradable materials are preferred over viral systems. Non-viral delivery systems permit the molecular design, at least in principle, of a vehicle with controlled size and architecture that can properly accommodate the desired transgene for a specific application37-40. The non-viral systems mentioned above decrease enzymatic degradation of chemically unmodified oligonucleotides and partially increase their cellular uptake. However, these systems are generally difficult to use and they show extracellular non-specific interactions in vivo, have problems in intracellular trafficking to the nucleus and mostly do not work in the serum containing media41. Furthermore, highly positively charged polymers such as polyethylenimine (PEI) cause cellular toxicity42. Therefore, new systems are needed to improve cellular uptake and reduce side effects.
6
1.2 Use of Peptide Amphiphile Nanofibers in Medicine
Peptide amphiphiles consist of a hydrophilic and hydrophobic group. Hydrophilic peptide headgroup is made up of charged amino acid residues such as net negative (aspartic acid (D), glutamic acid (E)), net positive (histidine (H), lysine (K), arginine (R)), or a mixture of positive and negative residues while hydrophobic tails consist of 3–9 non-polar amino acid residues (e.g. glycine (G), alanine (A), valine (V), phenylalanine (F), proline (P), isoleucine (I), leucine (L)) or alkyl tail that are composed of hydrocarbon chains of 12–16 carbons or mixture of hydrocarbon chain and non-polar amino acids43. During molecular self-assembly of peptide amphiphiles, amino acids could be spontaneously organized into larger well ordered nanostructures using non-covalent bonds; hydrophobic interactions, electrostatic forces, hydrogen bonding and aromatic interactions44. The morphology of the final assembled structure depends on the structure of the monomer, peptide length and composition of peptide amphiphiles. Peptide sequence that carries specific information on their side chains such as hydrophilicity, hydrophobicity and chirality and composition of peptide amphiphiles such as length of the hydrophobic part determine self assembly process. Self Assembly process results in different final nanostructures since formation of β-sheet and α-helix secondary structures depend on hydrophobic and hydrophilic residues such as the geometry of the polar head group and the shape of each molecule45. Several self-assembling peptidic aggregates have been designed including nanotubes, nanovesicles, nanobelts
7
and nanofibers with hydrophobic tail collapsed inside the core of the structure and hydrophilic residues projected on the outside, facing water46
Peptide amphiphiles (PA), which have an alkyl carbon tail linked covalently to the N terminal of a peptide sequence named lipopeptides, were developed by Hartgerink et al47. These PAs are composed of a hydrophobic site, a β-sheet forming short peptide sequence, a hydrophilic peptide sequence, and a bioactive peptide group. These peptide amphiphiles self assemble through weak, noncovalent interactions such as intermolecular association of the hydrophobic portions with the help of intermolecular polar interactions; electrostatic forces, hydrogen bonding and aromatic interactions.
PA molecules form highly ordered, one-dimensional cylindrical nanostructures through self-assembly. Hydrophobic site of PA molecules, composed of a long alkyl chain, induces hydrophobic collapse and remains in the interior part of the nanofiber structure while the hydrophilic peptide segment with net charge enables solubility in water and presents bioactive peptide sequence to the surface of these self-assembled nanostructures48-49. The β-sheet structures are formed through intermolecular hydrogen bonding between peptides, oriented parallel along the axis of the well-defined, one-dimensional nanofiber structures around 8-10 nm in diameter and up to a few micrometers in length depending on composition of peptide sequences47, 49-51
. Transition of the amphiphilic self-assembly of spherical micelles to cylindrical nanofibers is promoted by beta sheet formation with alanine, valine, glycine, and leucine residues that drive hydrogen bonding among
8
peptide segments. These self-assembled materials are sensitive to environmental changes such as temperature, pH, and ionic strength of the solution, as well as the concentration of the monomer52. Neutralization of the charged groups by addition of oppositely charged molecules such as biomacromolecules (e.g. DNA, protein, polysaccharides), divalent ions or change in pH trigger spontaneous nanofiber assembly of PAs through charge screening with the help of hydrogen bond formation. Water absorption by the mesh-like network formed by bundles of these nanofibers results in gel formation53-54. Chemical, physical and biological properties of nanofibers can be controlled by non-covalent bonds and supramolecular interactions55.
Peptide amphiphiles (PA) are promising tools for drug and gene delivery systems since they are biocompatible and bioactive. Gels formed by PAs have been used for various applications in regenerative medicine as extracellular matrix (ECM) – mimicking materials for repairing damaged tissues56, in cancer studies to kill cancer cells with anti-cancer epitope57, and in drug and gene delivery to improve stability and transfection efficacy. PA gels are comprised of meshwork of nanofibers with biofunctional epitopes derived from extracellular matrix proteins to form synthetic matrices that mimic natural ECM and have been used in regenerative medicine applications; such as angiogenesis induction58, neural59-60, bone61, cartilage regeneration62 and survival of pancreatic islets63. In gene delivery applications, PAs that are composed of hydrophobic amino acids or alkyl tail and hydrophilic group is functionalized with bioactive sequences and cell penetration peptide sequences to facilitate cellular uptake. RGDS sequence
9
derived ECM protein; fibronectin, provides recognition system with integrin receptors in cell surfaces. The integrins constitute a family of cell-adhesion receptors that simultaneously interact with the proteins in the extracellular matrix and the internal constituents of the cell and it is known that expression of integrins is increased in cancer cells. By this way, integrins provide communication between cell and its environment, allowing the cell to respond to its environment and regulate cell morphology, differentiation, proliferation, and gene expression64-65. Cell penetration peptide sequences such as, penetratin derived from Antennapediaprotein (AntpHD) and Tat derived from human immunodeficiency virus 1(HIV1) trans-activating transcriptional activator; enable the intracellular delivery of polar, biologically active compounds in vitro and in vivo65, 66-70.
PAs were also functionalized for slow release of proteins such as sonic hedgehog (SHH) protein71, and to deliver or encapsulate growth factors (e.g. bone morphogenetic protein factor-272 and fibroblast growth factor73). Koutsopoulos and his colleagues used hydrogels of the ac-(RADA)4-CONH2 peptide monomer for controlled release of functional proteins; insulin, trypsin inhibitor, BSA, and IgG through nanofiber74. Moreover PA nanofibers were employed to deliver and encapsulate hydrophobic drugs including pyrene75, doxorubicin76, cisplatin77, and ellipticine78.
10
3.1 Peptide Amphiphiles for Release and Delivery of Antisense ODN
Herein, we evaluate self-assembled nanofiber formation of PA system with G3139 oligonucleotide as a slow release and delivery system. We demonstrate a new carrier system to deliver G3139 as an antisense oligonucleotide drug model. G3139 (Genasense®, Genta Inc., NJ) is a chemically modified 18-mer antisense oligodeoxynucleotide (ODN) which is modified chemically with phosphorothioate, designed to bind to the first six initiation codons of the Bcl-2 mRNA. Heteroduplex of mRNA and ODN activates RNase H, which cleaves the mRNA sequence, resulting in the degradation of mRNA, thereby inhibiting the production of Bcl-2 protein ( Fig. 2)79-80. Bcl-2 protein family is crucial for mediating controlled cell death and overexpression of Bcl-2 protein leads to accumulation of aged and damaged cells resulting in tumor formation. Although Bcl-2 was discovered first with chromosomal translocation of t (14; 18), which occurs in the majority of indolent B-cell non-Hodgkin lymphomas, it has been associated with many other aggressive tumor phenotypes such as melanoma, lymphoma, lung, colon, prostate cancers and breast cancer81- 84.
G3139 has been shown to effectively reduce both Bcl-2 mRNA and protein expression and enhances the chemotherapeutic effects of a broad range of chemotherapy agents, including chemotherapy, radiation, monoclonal antibodies and immunotherapy in preclinical studies85-91. G3139 has been in clinical trials since 1995 in the U.S.A., Europe and Australia, with efficacy and safety data from phase 1, phase 2 and phase 3 clinical
11
trials. G3139 is now being evaluated clinically in the treatment of melanoma, chronic lymphocytic leukemia, lymphoma, acute myelogenous leukemia, advanced merkel cell carcinoma, lung, colon, prostate cancers and breast cancer as monotherapy or in combination with many types of anticancer therapies82-84, 92-100
In this study, we used a cationic peptide amphiphile molecule, Lys-PA (C12-VVAGK-Am), as a non-viral antisense oligonucleotide carrier system. Positively charged carrier systems for oligonucleotide delivery have been used to provide effective cellular uptake through enhanced binding to negatively charged proteoglycans on the cell surface101-102. Besides, these peptide amphiphiles have good affinity to cell membranes since hydrophobic content of the PA molecules facilitates internalization through cell membrane lipids103. The hydrophilic lysine residues were chosen to bind to oligonucleotides and form nanofibers at physiological pH upon addition of oligonucleotides through charge screening. An amphiphilic peptide forming β-sheet structure with four non-polar amino acid residues and positively charged lysine residue at physiological conditions was designed and synthesized to interact with oligonucleotides. Poly-amine groups presented on the surface of the self-assembled nanofibers provide positive charge at physiological pH that can facilitate the uptake of Lys-PA - ODN complexes104-105. Oligonucleotides can be released by degradation of the biodegradable peptide scaffold or by physical release mechanisms governed by diffusion. Cells can uptake nanofibers degraded by proteases and use them in their metabolic pathways showing biocompatibility of PAs106.
12
RGDS-PA (C12-A3KKRGDS-Am) was the second peptide amphiphile in our study. This peptide amphiphile sequence is comprised of hydrophobic alkyl tail, three alanines and two lysines with RGDS (arginine-glycine-aspartic acid- serine) residues. Unlike Lys-PA, bioactive RGDS sequences were also incorporated into the peptide segments to improve the internalization pathway and enhance targeting cellular uptake through receptor-mediated endocytosis107. Three-alanine sequence and positively charged two lysine residues at physiological conditions were designed and synthesized to form β-sheet structure and interact with oligonucleotides, respectively.
Controlled release of ODN from Lys-PA - ODN system, ODN internalization by cells and effects of this nano-carrier system on Bcl-2 mRNA levels are demonstrated. We showed that Lys-PA and ODN molecules form nanofibers through charge screening by using circular dichroism, SEM and AFM imaging. Lys-PAs undergo self assembly with addition of ODN to form dynamic nanostructure and affinity of ODN to Lys-PA analyzed with FRAP experiment. We showed that Lys-PA - ODN assembly provides controlled release of ODN from peptide nanofibers, enhanced intracellular accumulation of ODN and downregulation of Bcl-2 mRNA. We also showed that incorporation of bioactive RGDS sequence to the peptide increased cellular uptake of ODN.
13
Figure 2. Mechanism of action for Genasense®. Reproduced with permission from Genta Inc., NJ
14
EXPERIMENTAL SECTION
2.1. Materials
9-Fluorenylmethoxycarbonyl (Fmoc) and tert-butoxycarbonyl (Boc) protected amino acids, [4-[α-(2',4'-dimethoxyphenyl)Fmoc-aminomethyl]phenoxy]acetamidonorleucyl-MBHA resin (Rink amide MBHA resin) and 2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from NovaBiochem and ABCR. DMEM media, fetal bovine serum (FBS), trypsin EDTA and penicillin streptomycin were purchased from Invitrogen.
2.2. Antisense Oligonucleotides
G3139, a phosphorothioate oligodeoxynucleotide (Genasense®), with a sequence complementary for the first six codons of the open reading frame of Bcl-2 mRNA: 5′-tct ccc agc gtg cgc cat-3′ (18-mer) was used as the antisense ODN. A two-base mismatched (MM) sequence, G4126, 5′-tct ccc agc atg tgc cat-3′, was used as control and G3139 labeled with 6-fluorescein on the 5‟-t, G4243, 5′-tac cgc gtg cga ccc tct-3′, was used in cell internalization assays. ODNs were donated by Genta Inc. (Genta Inc., NJ).
15 2.3. Cells
The human breast cancer cell line MCF-7 (American Type Culture Collection) was maintained in DMEM low glucose (GIBCO) supplemented with 10% FBS, 100 µg/ml streptomycin and 100 µg/ml penicillin at 37 °C in a humidified incubator with 5% CO2.
2.4. Peptide Synthesis, Purification and Characterization
2.4.1. Synthesis of Peptide Amphiphiles
Lauryl-VVAGK and Lauryl-AAAKKRGDS peptides were constructed on MBHA Rink Amide resin. Amino acid couplings were done with 2 equivalents of Fmoc protected amino acid, 1.95 equivalents of HBTU and 3 equivalents of DIEA for 2 h. Fmoc removal was performed with 20% piperidine/dimethylformamide (DMF) solution for 20 min. Cleavage of the peptides from the resin was carried out with a mixture of TFA: TIS: H2O in ratio of 95:2.5:2.5 for 3 h. Excess TFA was removed by rotary evaporation. The remaining viscous peptide solution was triturated with ice-cold ether and the resulting white product was freeze-dried. Peptide was characterized by liquid chromatography-mass spectrometry (LC-MS). Mass spectrum was obtained with Agilent LC-MS equipped with Zorbax SB-C8 4.6 mm x 100 mm column. A gradient of (a) water (0.1% formic acid) and (b) acetonitrile (0.1% formic acid) was used for LC-MS characterization. Peptide
16
purification was performed with a Zorbax prepHT 300SB-C8 column with water-acetonitrile (0.1% TFA) gradient.
2.4.2. Characterization of Peptide Amphiphiles
Circular Dichroism (CD) Spectra Measurement
JASCO J815 CD spectrapolarimeter was used at RT to analyze secondary structures in 400 µl of peptide solution alone at a final concentration of 3.8 x 10-4 M of Lys-PA at pH 7 or 0.09 x 10-4 M of RGDS-PA at pH 10 or pH 6 and mixture of Lys-RGDS-PA (3.8 x 10-4 M final) and ODN (3.125 µM final) solution or RGDS-PA (0.09 x 10-4
M final) and ODN (1 µM final) solution at wavelengths ranging from 260 nm to 190 nm, data interval and data pitch being 0.1 nm, scanning speed being 100 nm/min. All measurements were done with three accumulations. D.I.T. was selected as 1 sec, band width as 1 mm, and the sensitivity was standard.
Viscoelasticity and Gelation Behavior
Gelation behaviors of the ODN and Lys-PA gels were evaluated by an Anton Paar Physica RM301 Rheometer operating with a 25 mm parallel plate configuration at 25 °C. Each sample with 100 µL of total volume with a final peptide concentration of 1 wt% and ODN solutions with different concentrations was carefully loaded on the center of the lower plate and incubated for 15 min before performing rheometry analysis. After
17
equilibration, the upper plate was lowered to a gap distance of 0.5 mm. Storage moduli (G′) and loss moduli (G″) values were measured from 100 rad/s to 0.1 rad/s of angular frequency with 0.5% shear strain.
Morphological Observation
Three-dimensional network of the nanofibrous gel was observed by atomic force microscopy (AFM) and scanning electron microscopy (SEM).
Atomic Force Microscopy (AFM)
AFM sample was prepared by mixing 0.02 wt % Lys-PA or 1 wt % RGDS-PA and ODN solutions in ddH2O. After incubation for 30 min at RT, mixture of Lys-PA and ODN was diluted 20 times in H2O or mixture of RGDS-PA and ODN was diluted 100 times to obtain 0.5 µM ODN concentration. Final solution was drop-casted and dried on a freshly cleaved silicon wafer. AFM images were recorded using model MFP-30 from Asylum Research operated in tapping mode at a frequency of 246 kHz. AFM images were taken at 1024×512 pixels resolution. Image was taken with spring constant 40 N/m and the set point and scanning speed were 0.7–1.0 V and 1.0–1.5 Hz, respectively.
18 Scanning Electron Microscopy (SEM)
3-D network of self-assembled 100 μl gels prepared on a metal mesh by mixing 50 μl of 1 wt% PA solution in H2O with 50 μl of ODN solution in PBS (1X, pH 7.4) containing 250 ng/µl of ODN. Water was exchanged with ethanol gradually in water/ethanol mixtures of increasing ethanol concentrations and finally in 100% ethanol for 30 sec at each step. Gel was dried at critical point (1072 psi, 31 ºC) with Tousimis Autosamdri-815B, Series C critical point dryer and coated with 10 nm Au-Pd. Sample was imaged by a FEI Quanta 200 FEG, using the ETD detector at high vacuum mode at a voltage of 30 keV
2.5. ODN Release from PA-ODN Gel
ODN release from PA-ODN gel was examined by diluting Lys-PA with H2O to 2 wt% in 50 µl and mixing with 50 µl ODN solution (30 µg/µl and 2 µg/µl) in PBS (1X, GIBCO) to obtain a final 1 wt% peptide gel and 15 µg/µl ODN in the gel. After 15 min incubation, 100 µL of 1X PBS was added onto gel. Release of ODN was measured by NanoDrop (The Thermo Scientific NanoDrop 2000, USA) at 260 nm as initial release and measurements were continued for a period of 6 days for gel formed with 30 µg/µl ODN and 3 days for gel formed with 2 µg/µl ODN solution. To examine the effect of PA and ODN concentration on release profile of ODN; 100 µl of gels were formed using ODN with concentrations of 150-1200 ng/µl and PA with
19
concentrations of 0.2-0.1 wt%. The gels were prepared in 96 well-plates and incubated for 1 h at 37 ºC followed by overnight drying in a laminar flow hood. Next day, 100 µl of PBS (1X) was added on coated wells and gels were maintained at 37 ºC for a period of 5 days during ODN concentration analysis. To see burst release, 2 µl aliquots from PBS solution were measured in short time intervals in the first day and every 24 h afterwards. 2 µl of fresh buffer was added to wells after each measurement for all experiments. ODN release versus time was graphed according to initial concentration of ODN trapped in the gel. Three replicates were tested for each PA and ODN formulation.
2.6. Confocal Fluorescence Recovery after Photobleaching of FAM-Labeled ODN in PA-ODN nanofibrous network
For photobleaching experiments, fluorescent gels were obtained by mixing 10 µL of 0.1 wt % peptide amphiphile solution and 10 µL of 1000 ng/µL oligonucleotide solution (97% ODN and 3% FAM-ODN) after 10 min incubation at RT. Fluorescence images of the samples were acquired using the laser scan confocal microscope (ZEISS LSM 510 META, ZEISS GmbH, Jena, Germany) with a 63×, 1.2 NA oil-immersion objective using the 488 nm line of an argon laser and the 500–530 nm band pass filter at a 512 pixel × 512 pixel resolution, at a scan speed of 9 and using a 95 μm pinhole. FRAP experiments were carried out by scanning a square region of interest (ROI) and bleaching a circular ROI by exposing to 100 % 488 nm
20
light. Recovery of fluorescence in this area and unbleached area was obtained by a time series using a low laser intensity of 0.5 % 488 nm light.
2.7. In Vitro Studies
2.7.1. Cellular Uptake of PA-ODN Complexes
MCF-7 breast cancer cells (3×104 cells/ well) were plated in 0.5 ml of growth medium on coverslips in 24-well chambered plates and grown overnight. On the following day, the cells were treated with 20 µl of mixture with an equal volume of Lys-PA and FAM-labeled ODN (G3139) or 10 µl of FAM labeled ODN alone for 4 h. Mixture of 0.01 wt% Lys-PA and FAM labeled ODN (30 ng/µl) in PBS and or FAM labeled ODN (120 ng/µl) in PBS and 1 wt% RGDS-PA in H2O was prepared by gently mixing ODNs and PAs, followed by incubation for 30 min at RT. After 4 h incubation, cells were rinsed three times with PBS, fixed with 4% paraformaldehyde and stained with 1 µg/ml solution of TO-PRO-3 (Invitrogen) for nuclei staining. In order to see distribution of PA-ODN complex in cell cytoplasm, after 4 h incubation with FAM labeled ODN alone and in PA-ODN mixture, cell culture medium was removed, 500 μl/well fresh cell culture medium was added and cells were incubated for an additional 44 h. Cells were imaged by a laser scan confocal microscope (ZEISS LSM 510 META, ZEISS GmbH, Jena, Germany).
21 2.7.2. Cell Viability and Proliferation Assay
The cytotoxicity of the ODN, PA and PA-ODN complexes was measured by MTT [3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay (Sigma). The MCF-7 cells were plated on 96- well plates at a density of 3000 cells/well and grown overnight. Next day, the cells were treated with either a 5 µl mixture of equal volumes of Lys-PA (0.02 wt%) and ODN/MM-ODN or a 2.5 µL solution of ODN/MM-ODN alone with final concentration of 1 µM .The cells were treated with either a 5 µl mixture of equal volumes of RGDS-PA (1 wt %) and ODN/MM-ODN or a 2.5 µL solution of ODN/MM-ODN alone with final concentration of 1 µM. After incubation at 37 °C for 4 h, cell culture medium was replaced with fresh medium followed by additional 44 h incubation.MTT assay was performed according to the manufacturer's protocol. Briefly, media on cells were aspirated and 100 µl fresh media containing 10% (v/v) reconstituted MTT (M-5655) reagent was added to each well. The plates were incubated for 3 h and the absorbance of the medium was measured at 570 nm with plate reader (SpectraMax M5, Molecular Devices). The background absorbance of multiwell plates were also measured at 690 nm and subtracted from the 570 nm measurement.
To confirm the effect of PA-ODN complex on proliferation of MCF-7 cells, BrdU cell proliferation kit (Roche) was used. Cells were seeded at a density of 3000 cells/well in a final volume of 100 μl in 96-well culture
22
dishes and incubated overnight. The cells were treated with either a 5 µl mixture of equal volumes of Lys-PA (0.02 wt %) and ODN/MM-ODN or a 2.5 µL solution of ODN/MM-ODN alone with final concentration of 1 µM . After incubation at 37 °C for 4 h, cell culture medium was replaced with fresh medium followed by additional 20 h incubation. . Next, 10 μl of 100 µM BrdU label was added to each well and incubated for an additional 24 h at 37 °C. The labeling medium was removed by tapping off and replaced with 200 μl of fix/denaturing solution followed by 30 min incubation at RT. The fix/denaturing solution was removed thoroughly by flicking off and tapping. 100 µl/ well anti-BrdU-POD working solution was added to each well and left for 90 min at RT. Antibody conjugate was removed by flicking off and wells were rinsed 3 times with 200 -300 µl/well with washing solution (PBS, 1X). Finally 100 µl/well of substrate solution was added into wells and incubated at RT until color development is sufficient for photometric detection (30 min). Absorbance (A370 nm - A492 nm) of the samples was measured with plate reader (SpectraMax M5, Molecular Devices). Absorbance values were normalized with respect to cells incubated without ODN drug. Assays were carried out in triplicate.
2.7.3. Determination of Bcl-2 mRNA Expression by Quantitative RT-PCR
MCF-7 breast cancer cells were plated in 6-well plates containing 2 ml media. Following overnight growth at 37 ºC in a humidified incubator with 5% CO2, cells were treated with either a 100 µl mixture of equal volumes of
23
Lys-PA (0.02 wt %) and ODN or a 50 µL solution of ODN/MM-ODN alone with final concentration of 1 µM. After incubation at 37 °C for 4 h, cell culture medium was replaced with fresh medium followed by additional 44 h incubation.. The complexes of PA-ODN were prepared by mixing equal volumes of ODN solution in PBS (1X, GIBCO) and Lys-PA (0.02 wt %) in ddH2O and incubated for 30 min at RT before adding of this mixture onto cells. Total RNA was extracted from cells after 48 h and 72 h incubations by using Trizol reagent (Invitrogen). RNA concentration was measured with NanoDrop spectrophotometer (Thermo Scientific NanoDrop 2000, USA) at 260 nm.
The Bcl-2 mRNA expression level was measured by using quantitative RT-PCR with SuperScript™ III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen). Samples were incubated in C1000TM thermal cycler (CFX96TM real Time systems, Bio-RAD). After cDNA synthesis for 3 min at 50 ºC, cDNA denaturation was carried out for 5 min at 95 ºC followed by denaturation at 95 ºC for 15 sec and annealing at 60 ºC for 30 sec. Primer sequences were designed by using DNASTAR Lasergene 8 program. Following primer pairs were used for qRT-PCR experiments: 5‟tgc ccc tgt gga tga ctg ag‟3 and 5‟gtt tgg ggc agg cat gtt gac t‟3 for Bcl-2, 5‟tcg aca gtc agc cgc atc ttc t‟3 and 5‟gtg acc agg cgc cca ata cga c‟3 for GAPDH. Relative gene expression values were determined by the Pfaffl method. GAPDH was used as housekeeping gene to normalize the gene expression levels of each sample.
24 2.7.4. Western Analysis
MCF-7 breast cancer cells were plated in 6 well plates containing 2 ml media. Following overnight growth at 37 °C in a humidified incubator with 5% CO2, cells were treated with either a 100 µl mixture of equal volumes of Lys-PA (0.02 wt%) and ODN or a 50 µL solution of ODN/MM-ODN alone with final concentration of 1 µM . After incubation at 37°C for 4 h, cell culture medium was replaced with fresh medium followed by additional 44h and 68 h incubation. The complexes of PA-ODN were prepared by mixing equal volumes of ODN solution in PBS (1X, GIBCO) and Lys-PA (0.02 wt %) in ddH2O and incubated for 30 min at RT before adding of this mixture onto cells. After 48 h and 72 h incubation, MCF-7 cells were washed with 1X PBS and lyzed with 100 μl 1X SDS-sample buffer. The extract was transferred to a microcentrifuge tube and centrifuged at 13,000 x g for 10 min at 4 ºC. The supernatant was collected to a fresh tube and the total protein concentration was determined by using BCA protein assay (Pierce, Rockford, IL). Cell lysates containing 50 μg of total protein were loaded and separated on a 10% SDS-polyacrylamide gel at 150 V for 1.5 h. After blotting (trans-blot SD, Semi Dry transfer cell, Bio-Rad) to polyvinylidine difluoride membranes (Thermo Scientific) at 14 V for 35 min, the membrane was blocked in a solution of 5% powdered nonfat milk in Tris-buffered saline/Tween-20 followed by incubation with monoclonal anti-Bcl-2 clone 100 (Millipore) (1:1000) or monoclonal Anti-β-Actin antibody (1:5000) (Sigma-Aldrich). After washing with milk, the
25
membranes were incubated with horseradish peroxidase conjugated goat anti-mouse secondary antibody (Millipore) (1:2000). The membranes were developed with Novex® Chemiluminescent Substrates-(Invitrogen) and observed with Kodak Biomax NS film.
26
RESULTS AND DISCUSSION
3.1. Peptide Synthesis, Purification and Characterization
Lauryl-VVAGK-Am (Lys-PA) (Fig. 4a) peptide was synthesized by utilizing solid phase peptide synthesis method (Fig. 3). The mass spectrometry and HPLC chromatogram are shown in Figures 4 and 5. Lauric acid provides amphiphilicity with its hydrophobic character that triggers peptide molecule to self assemble into nanofibers. The β-sheet forming group consists of four non-polar amino acid residues to form β-sheet module. Lysine residue provides water solubility and a cationic feature which triggers self supporting gel formation upon mixing with negatively charged oligonucleotides (Fig. 4c) through self assembly process (Fig. 7).
Secondary structures of the nanofibers formed through charge neutralization of the Lys-PA molecules by mixing with ODNs were analyzed by circular dichroism (CD). Circular dichroism examination of PA with ODN demonstrated that PA formed predominantly β-sheet structure at a concentration of 3.8 x 10-4 M in water. The typical β-sheets display a negative minimum at 218 nmand a positive ellipticity at 195 nm suggesting the b-sheet contents and b-sheet packing, respectively108. CD spectrum of PA without ODN at the same concentration revealed a mixture of random coil and β-sheet structures109. The amount of β-sheet structures in the PA
27
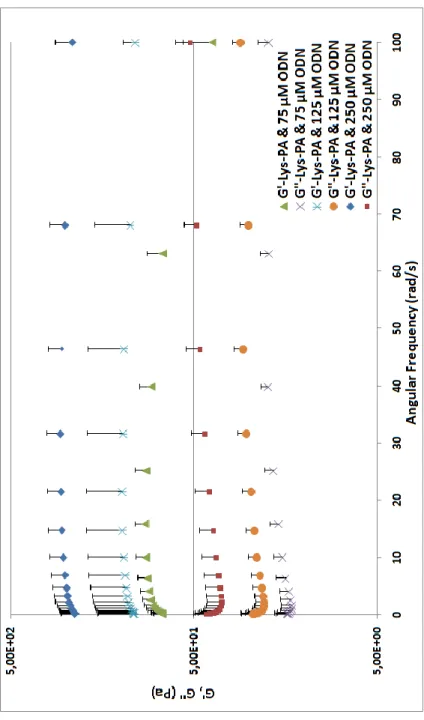
alone samples was much lower compared to CD spectrum of the PA with ODN (Fig. 8). Characterization of PA-ODN solution by AFM and gel by SEM imaging revealed their 3-D nanofibrous network structure (Fig. 9a and b). Lys-PA, which is positively charged at acidic pH, can form gel through neutralization by making the pH basic or by addition of positively charged ODN at physiological pH. Lys-PAs self-assemble into nanofibers upon addition of oppositely charged ODN through charge screening and three dimensional network of these nanofibers results in gel formation upon water encapsulation. This observation suggests that nonporous structure can be used for controlled release of DNA based drugs and therapeutic proteins. Network formation is critical for drug release since it enables physical encapsulation of the drug in addition to electrostatic interactions between the ODN and PA molecules. In addition, ODN release is dependent on electrostatic interactions between the PAs and ODNs. Therefore, PA degradation can be responsible for additional ODN release in vitro and in vivo. Encapsulation of water by the 3-D nanofibrous network resulted in gel formation triggered by addition of ODN solution as demonstrated by oscillatory rheology. G‟ and G” did not change with angular frequency from 100 rad/s to 0.1 rad/s (Fig. 10). As we increased the ODN amount, storage moduli (G‟) of samples increased and became significantly higher than their loss moduli (Fig. 11). G′/G′′ was higher than 1, a critical point of gelation, which indicates PA-ODN gel behaves like an elastic solid54.
28
Figure 3. Solid Phase Synthesis Diagram. Reproduced with permission from Sigma-Aldrich.
29
Figure 4. (a) Chemical representation of Lys-PA, (b) RGDS-PA and (c) Chemical structure of the backbone of the phosphorothioate G3139.
30
31
32
Figure 7. Schematic representation of the fibrous gel of Lys-PA (2 wt %) and Fluorescein tagged-ODN (1000 ng/µl)
33
Figure 8. CD spectra demonstrate increased β-sheet formation in PA- ODN mixture compared to PA alone
34
Figure 9. (a) AFM topography and (b) SEM images of nanofibrous 3-D network of the Lys-PA and ODN complex
35
Figure 10. Frequency sweep profiles of gels of Lys-PA (2 wt %) with ODN at different final concentrations (75-250 µM). Data points are average of n=3
36
Figure 11. Storage modulus (G′) and loss modulus (G′′) of gels of Lys-PA (2 wt %) with varying concentrations of ODN at angular frequency of 100 rad/s. Data points are averages of n=3 and error bars represent standard
37
3.2. ODN Release from PA-ODN Gel
We evaluated the effect of the Lys-PA and ODN concentrations on the release of ODN through release assays. This system offers a distinctive advantage that self-assembly process is triggered with addition of ODN or any other desired drug that can be directly encapsulated into network compared to the loading of the drugs. The nanofibrous nature of the PA-ODN system enables physical encapsulation of the PA-ODN as well as increasing the physical interaction between the PA and ODN molecules. The release profile of ODN from gel prepared by mixing 50 µl of 2 wt% Lys-PA and 50 µl of 30 µg/µl ODN is shown in Figure 12a. We monitored sustained release of ODN from 3-D PA-ODN nanofibrous network while the majority of ODN (80–90%) was released by 6 days (Fig. 12a). Effect of charge ratio is an important factor that determines the strength of the electrostatic interactions and it was further studied by changing peptide concentration in the PA-ODN gels. For this purpose, ODN release was studied with a gel of 2 µg/µl (2000 ng/µl) ODN and 2 wt% Lys-PA for 3 days (Fig. 12b). Representative image of PA-ODN gel is shown in Figure 12c. Release of ODN from gel formed with 30 µg/µl ODN solution was around 55% by day 3 while release of ODN from gel formed with 2 µg/µl ODN was around 15% with a slower release profile. In addition, figure 13 demonstrates that ODN release from gel decreased as the peptide concentration was increased due to enhanced attraction of anionic ODNs by the cationic nanofibrous network. The average release profile was also affected by the concentration of ODN
38
in the gel, where higher the ODN concentration, higher the ODN release from gel due to insufficient attraction by cationic Lys-PAs on anionic ODNs in the gel. At low ODN concentrations (150 ng/µl in Fig. 13a and 300 ng/µl in Fig. 13b), 66% of the initial amount of ODN was trapped in the 0.1 and 0.2 wt% Lys-PA-ODN gels and release from the 3-D network was slow for all three concentrations of PAs. Release assay from the gels with 600 ng/µl ODN concentration showed rapid initial release within the first 3 h when 0.1 wt% gel was used where slower release profiles from 0.15 wt% and 0.2 wt% gels (Fig. 13c). When a higher concentration (1200 ng/µl) of ODN was used for release assays, rapid initial release within the first 3 h was observed for all three PA concentrations (Fig. 13d). After 2 days, release profile reached plateau values for all formulations. When same PA concentration was used for encapsulating different concentrations of ODN, using lower concentrations of ODN resulted in slower release while as ODN concentrations were increased, more rapid initial release profiles were observed.
Antisense oligonucleotides are not only localized on the surface of the nanofibers by electrostatic interactions between positively charged PA molecules and negatively charged ODNs, but also physically encapsulated inside the peptide nanofiber network. The initial burst release is due to weaker interactions between the ODNs and the PAs which is caused by physical encapsulation. The latter slow release is likely to be caused by stronger bonds caused by electrostatic interactions between the PA and ODN molecules. Therefore, self-assembled PA and ODN nanofibers can be used
39
for controlled ODN release by changing PA/ODN charge ratio and the PA-ODN nanofiber network provides a promising tool for controlled release of oligonucleotides. It is known that oligonucleotides have short half-lives in the body. Consequently, agent-free antisense ODN (G3139) is generally applied by continuous intravenous infusion. Thus, utilization of an effective delivery system is required to improve the efficacy of gene targeted therapy while providing elimination of side effects in the circulatory system after injection.
The fluorescein labeled ODN in the PA-ODN gel was photobleached in an area 173 µm2 and the recovery of fluorescence was monitored over time in the same area and compared with fluorescein intensity of non-bleached area. After photobleaching of fluorescein-ODN irreversibly, mobile/unbound fluorescent ODN or under dynamic assembly process, unbleached fluorescein-ODN molecules moved to the bleached area and caused recovery of the fluorescein. Recovery occurred almost after 8 minutes suggesting that ODN has high affinity to PA (Fig. 14). Although determination of the diffusion constant of ODN in the PA network is difficult by FRAP experiment, affinity of different additives such as other peptides or macromolecules to peptide amphiphiles can be compared.
40
Figure 12. Release profile shows of ODN from peptide nanofiber (a) gel of 30 µg/µl ODN and 2 wt % PA for 6 days, (b) gel of 0.2 µg/µl ODN and 2 wt % PA
for 3 days. (c) Representative image of gel with PBS used for release experiments. Data points are average of n=3 and error bars represent standard
41
Figure 13. Release profile of ODN from (a) gel of 0.1%, 0.15% and 0.2 wt% PA and ODN at concentrations of 150 ng/µl, (b) 300 ng/µl, (c) 600 ng/µl, and
(d) 1200 ng/µl for 5 days. Data points are average of n=3 and error bars represent standard error of mean.
42
Figure 14. Intensity graph of area of fluorescein ODN area at time of pre-bleach, post bleach and post-recovery compared to unbleached area
43 3.3. In Vitro Studies
Incorporating ODNs into self-assembled Lys-PA nanofibers also enhanced ODNs‟ in vitro activity and their cellular uptake. Intracellular distribution of PA-ODN complexes was examined by laser scan confocal microscopy (Fig. 15a). MCF-7 cells were incubated with FAM labeled PA-ODN complex and free FAM labeled PA-ODN for 4 h at 37°C followed by fixation. In order to see the distribution of PA-ODN complexes in cell cytoplasm over time, cells were treated with PA-ODN containing media for 4 h, after which the medium was removed. Fresh cell culture medium was added and cells were incubated for an additional 44 h before fixation. After 48 h, we observed that PA-ODN complexes were internalized by cells (Fig. 15b). The internalization of ODN by cells is continuous and thus effectiveness of ODN is sustained. Stronger fluorescent intensity was observed in the cells treated with PA-ODN complex after both 4 h incubation and 48 h incubation with respect to the ODN alone, indicating enhanced cellular uptake of ODN when it is applied with positively charged PA due to their amphiphilic and cationic properties.
In our case, ionic interactions of positively charged lysine residues with membrane-bound heparan sulfate proteoglycans (HSPGs) induce non-specific uptake of complexes. The exact mechanism of cellular internalization of ODN with peptide amphiphile nanofibers remains unclear and will be studied. The most likely way of internalization can be macropinocytosis formed by actin-driven membrane ruffling, by which
44
positively charged complexes are endocytosed in fluid-filled vesicles. These vesicles may fuse with degradative late endosomes or bypass the lysosomal compartment and traffic directly to the nucleus110-111. The proposed mechanism is represented in Figure 16.
In order to analyze the effects of Lys-PA - ODN complex on cell viability and proliferation, MTT viability and BrdU proliferation assays were carried out by using MCF-7 cells. Cells incubated with Lys-PA - ODN complex had lower viability than ODN alone, which proves that ODN is an effective delivery agent. Cells treated with Lys-PA - ODN complex also had lower viability compared to cells treated with Lys-PA - MM ODN complex, which shows that the decreased viability is not caused by the Lys-PA molecules. Proliferation assays revealed no significant differences between cells treated with ODN, MM-ODN, Lys-PA - ODN and Lys-PA - MM ODN. This result is consistent with previously published data112 (Fig. 17a and b).
Functional delivery of ODN with Lys-PAs was also confirmed by real time PCR analysis, which revealed that Lys-PA and ODN (G3139) complex induced downregulation of the Bcl-2 mRNA. MCF-7 cells treated with PA-ODN complex (G3139) exhibited a 70% downregulation in Bcl-2 mRNA levels when compared to treatment groups with Lys-PA - mismatch ODN (G4126), ODN (G3139) alone and mismatch ODN (G4126) alone when 4 h long treatment was applied to cells followed by 44 h of incubation without treatment (Fig. 18a). At 72 h, downregulation of Bcl-2 mRNA levels was
45
around 30% for cells treated with the Lys-PA - ODN complex, which was not significantly different than the other treatment groups (Fig. 18b). The increase in Bcl-2 mRNA levels at 72 h compared to 48 h might be caused by development of resistance by MCF-7 cells against ODN treatment, similar to previously published data113. All expression levels were normalized to cells without any ODN treatment. These findings suggest that PAs can effectively deliver ODNs for gene-targeted therapies.
The effect of the Bcl-2 antisense ODN (G3139) and its two base mismatch control (G4126) as naked and with Lys-PA on Bcl-2 protein levels in MCF-7 cells were also examined with western blot analysis, however, Lys-PA - ODN complex did not reveal any significant differences in the Bcl-2 protein levels compared to cells incubated with only cell culture medium. This result might be caused by the post-transcriptional and post-translational regulation mechanisms that are normally employed by cells to regulate protein levels and is a matter of effectiveness of G3139 itself which is also shown with QRT-PCR results. At 48 h (Fig. 19) and 72 h (Fig. 20), downregulation of Bcl-2 protein levels in cells treated with Lys-PA - ODN complex was not significantly different than the other treatment groups. Nevertheless, the reduction in Bcl-2 mRNA levels by itself at 48 h is sufficient to deduce that our delivery system enables effective delivery of the ODN agent. All in vitro experiments were carried out with serum containing media in which most of the ODN carrier systems do not work42. ODNs in the Lys-PA - ODN complexes are released by degradation of the biodegradable peptide scaffold. Synthesis and application of PAs as carrier
46
and delivery agents have advantage of convenience in usage over other biodegradable lipids and polymers. Moreover peptide amphiphile assembly occurs at physiological conditions spontaneously with addition of drug and injectable through small needles while polymers need addition of catalysts or material processing for the supramolecular self-assembly and reassembly event. Due to their biocompatibility, bioactive sequences and highly ordered nanostructures, PA nanofibers are promising drug and gene delivery systems.
47
Figure 15. Representative images of MCF-7 cells treated with (a) naked FAM labeled ODN (green); (b) FAM labeled ODN and 0.01 wt% PA complex for 4 h
followed by an additional incubation of 44 h without treatment at 37 oC. TO-PRO-3® (blue) was used to stain the nuclei. Images were taken at 63X
48
Figure 16. The proposed mechanism of PA – ODN complex (adapted from with permission from Leong et. al.)111
49
Figure 17. (a) Viability and (b) proliferation assay results of MCF-7 cell line incubated with naked ODN/MM (mismatch) and complex with Lys-PA. Data points are averages of n=3 and error bars represent standard error of mean.
50
Figure 18. Change in gene expression of Bcl-2 in MCF-7 breast cancer cells incubated with PA-ODN (G3139)/mismatch (G4126) complexes and free ODN/mismatch. Total RNA was extracted from cells after 4 h of treatment followed by (a) 44 h (b) 68 h of incubation without treatment. GAPDH was used
as housekeeping gene to normalize the gene expression level using Pfaffl method. Data points are averages of n=3 and error bars represent standard error
51
Figure 19. Western blot analysis of Bcl-2 protein expression in MCF-7 cells incubated for 48 h with Lys-PA - ODN complex.
Figure 20. Western blot analysis of Bcl-2 protein expression in MCF-7 cells incubated for 72 h with Lys-PA - ODN complex.
52 3.4. Future Perspectives
Lauryl-AAAKKRGDS-Am (RGDS-PA) (Fig. 4b) peptide was synthesized by solid phase peptide synthesis method (Fig. 3). The mass spectrometry and HPLC chromatogram of the molecule are shown in Figures 21 and 22. Lauric acid provides amphiphilicity with its hydrophobic character that triggers peptide molecules to self assemble into spherical aggregations in water (Fig. 23). Nonpolar amino acid residues of three alanine amino acids were used to form the β-sheet as secondary structure. However, CD spectra revealed that when RGDS-PA was mixed with ODN, random coil formation was the main aggregation form (Fig. 24). Due to the neutralization of head groups by electrolytes, the CD spectrum of the RGDS-PA solution at pH 10 shows the same tendency with RGDS-PA solution mixed with negatively charged ODN solution. In Figure 24, CD spectra of RGDS-PA at pH 10 and in mixture of ODN reveal random coil conformation with a negative minimum at 195 nm and a positive ellipticity at 212 nm109. On the other hand at pH 6, this PA did not display the typical random coil conformation that was evidence of a more unstable structure than the former. High pH causes neutralization of charges of head group and addition of ODN causes reduced hydrophilic head group repulsion. These result in formation of more compact RGDS-PA with spherical structures under charge screening. Furthermore, characterization of mixture of RGDS-PA with ODN by AFM imaging revealed that the RGDS-RGDS-PAs aggregate in spherical form but not in nanofibers (Fig. 25). We used three alanine