ContentslistsavailableatScienceDirect
Colloids
and
Surfaces
B:
Biointerfaces
jou rn a l h om ep ag e :w w w . e l s e v i e r . c o m / l o c a t e / c o l s u r f b
Bacteria
encapsulated
electrospun
nanofibrous
webs
for
remediation
of
methylene
blue
dye
in
water
Omer
Faruk
Sarioglu
a,b,
Nalan
Oya
San
Keskin
b,c,d,
Asli
Celebioglu
a,b,
Turgay
Tekinay
d,e,∗∗,
Tamer
Uyar
a,b,∗aInstituteofMaterialsScience&Nanotechnology,BilkentUniversity,06800,Bilkent,Ankara,Turkey bUNAM-NationalNanotechnologyResearchCenter,BilkentUniversity,06800,Bilkent,Ankara,Turkey cPolatlıScienceandLiteratureFaculty,BiologyDepartment,GaziUniversity,06900,Polatlı,Ankara,Turkey dLifeSciencesApplicationandResearchCenter,GaziUniversity,06830,Golbasi,Ankara,Turkey
eDepartmentofMedicalBiologyandGenetics,FacultyofMedicine,GaziUniversity,06500,Besevler,Ankara,Turkey
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received31August2016 Receivedinrevisedform 22December2016 Accepted18January2017 Availableonline19January2017
Keywords: Bacteria Bioremediation Electrospinning Encapsulation Methyleneblue Nanofibers
a
b
s
t
r
a
c
t
Inthisstudy,preparationandapplicationofnovelbiocompositematerialsthatwereproducedby encap-sulationofbacterialcellswithinelectrospunnanofibrouswebsaredescribed.Acommercialstrainof Pseudomonasaeruginosawhichhasmethyleneblue(MB)dyeremediationcapabilitywasselectedfor encapsulation,andpolyvinylalcohol(PVA)andpolyethyleneoxide(PEO)wereselectedasthepolymer matricesfortheelectrospinningofbacteriaencapsulatednanofibrouswebs.Encapsulationofbacterial cellswasmonitoredbyscanningelectronmicroscopy(SEM)andfluorescencemicroscopy,andthe via-bilityofencapsulatedbacteriawascheckedbylive/deadstainingandviablecellcountingassay.Both bacteria/PVAandbacteria/PEOwebshaveshownagreatpotentialforremediationofMB,yet bacte-ria/PEOwebhasshownhigherremovalperformancesthanbacteria/PVAweb,whichwasprobablydue tothedifferencesintheinitialviablebacterialcellsforthosetwosamples.Thebacteriaencapsulated electrospunnanofibrouswebswerestoredat4◦Cforthreemonthsandtheywerefoundaspotentially
storableforkeepingencapsulatedbacterialcellsalive.Overall,theresultssuggestthatelectrospun nanofi-brouswebsaresuitableplatformsforpreservationoflivingbacterialcellsandtheycanbeuseddirectly asastartinginoculumforbioremediationofwatersystems.
©2017ElsevierB.V.Allrightsreserved.
1. Introduction
Therearedifferenttypesofcontaminantsinwastewater efflu-entswhichareutilizedinindustrialprocesses,anddyescomprise agreat portioninthoseindustrial contaminants.Syntheticdyes haveagreatusageinvariousindustries(e.g.textile,leather,paper) anddyeingprocesscanleadtomanyenvironmentalproblems[1]. Methyleneblue(MB)isacommonbasic,cationicdyewithabroad applicationareaintextileindustry,paperindustry,chemistry,
biol-Abbreviations: MB,methyleneblue;PEO,polyethyleneoxide;PVA,polyvinyl alcohol.
∗ Correspondingauthorat:InstituteofMaterialsScience&Nanotechnology, BilkentUniversity,InstituteofMaterialsScience&Nanotechnology,Bilkent,06800, Ankara,Turkey.Tel.:+903122908987;fax:+903122664365.
∗∗ Correspondingauthorat:LifeSciencesApplicationandResearchCenter,Gazi University,06830,Golbasi,Ankara,Turkey.
E-mailaddresses:ttekinay@gazi.edu.tr(T.Tekinay),uyar@unam.bilkent.edu.tr
(T.Uyar).
ogyandmedicine[1,2–7].Nevertheless,decontaminationofMB fromwatersystemsafteruseisstillamajorchallenge. Conven-tionalwastewatertreatmentmethodscanbeusedforremediation ofMBsuchasphotocatalysis[8],advancedoxidationprocesses[9], reverse osmosis[10] and electrochemicaltreatment[11]. Some ofthesetechniquescanbeusedefficiently forMBremediation, thougheachofthemhastheirownlimitations,henceinnovative approaches have been presented in theliterature for develop-mentofsustainable,environmentallyfriendly,cost-effectiveand efficienttreatmentmethods[1].Bioremediationisanalternative technologyfordecontaminationofwatersystemsbyuseofspecific microorganisms,anditcanprovidegreen,efficient,cost-effective andsustainableremediationofwatercontaminants[12]. Microal-gae,fungiandbacteriacanbeutilizedforbioremediation.These microorganismscanremediatewaterpollutantseitherby biosorp-tionorbioaccumulation.Althoughdeadcellbiomassescanonlybe usedforbiosorption,livingcellscanpossessbothbioaccumulation andbiosorption,hencehigherefficiencyforbioremediationcould beachievedbylivingcellsinsomestudies[12].
http://dx.doi.org/10.1016/j.colsurfb.2017.01.034
246 O.F.Sariogluetal./ColloidsandSurfacesB:Biointerfaces152(2017)245–251
ThegenusPseudomonascomprisesGram-negative,aerobic, rod-shapedbacteriaandhasabroadmetabolicdiversity[13],havinga potentialtobeusedinbioremediationstudies.Apopular mem-berofthisgenus,Pseudomonasaeruginosahasalreadybeenused effectivelyinbioremediationoforganiccontaminants[1,14–16], implyingthisspeciesasapotentialcandidateforfurther bioreme-diationstudies.
Applicationofmicroorganismsforuseinbioremediationcan beperformedwitheitherfreemicroorganismsormicroorganism immobilized bio-hybridmaterials.Immobilized microorganisms can bring advantages than free cells in terms of their poten-tialreusability,lowerspaceandgrowthmediumnecessities,and higherresistancetoenvironmentalextremes[17,18].Electrospun fibrouswebshavebecomeapopularcarriermatrixfor immobi-lizationof specificmicroorganismsfor bioremediation of water systems[11,19–22].Electrospinningcanallowsimple,versatileand cost-effectiveproductionoffibrouswebswithuniqueproperties suchashighsurfaceareaandporosity,makingelectrospunfibrous websaspromisingcandidatesformicrobialintegrationand mem-brane/filterapplications[23].Inrecentyears,anumberofstudies havebeenreportedaboutencapsulationofmicroorganismsinto electrospunfibrousmatrices[23–27].Whileviabilityorbioactivity ofencapsulatedmicroorganismshasbeencheckedinallofthese studies,justonlyveryfewofthemhavereportedenvironmental applications[26–29].
In the present study, bioremediating bacterial cells were successfully encapsulated into polyvinyl alcohol (PVA) and polyethyleneoxide(PEO)polymericmatrices whilekeepingthe bacteriabioactiveandtheviablecellnumbersindesirableamounts. Two water based and biocompatible polymeric matrices were selectedforencapsulationofbacterialcellstoreducetheeffectsof exteriorenvironmentontheviablebacterialnumbers.Thesenewly producedbacteriaencapsulatednanofibrouswebsweretestedfor theirremovalcapacitiesagainstMBdye.Itwasfoundthat, bacte-riaencapsulatedwebshavethepotentialtosuccessfullyremediate MBinwater.Inaddition,thestorabilityofbacteriaencapsulated nanofibrouswebswastestedintermsoftheviablebacterial num-bers.Theresultshaveshownthattheencapsulatedbacteriacanbe storedsafelyforlongtimeperiodswithoutsignificantlossesintheir cellviability.Thesetypesofbio-hybridmaterialscouldbeof inter-estduetoeasyandsafepreservationofbioremediatingbacteriafor potentialwastewatertreatmentapplications.
2. Experimental
2.1. Materials
The chemicals and reagents (polyvinyl alcohol (PVA, Mw ∼125.000,Scientific PolymerProducts,Inc.),polyethyleneoxide (PEO,Mw∼900.000,Sigma-Aldrich),methyleneblue(MB,≥82%, Sigma-Aldrich),Nutrientbroth(Sigma-Aldrich),LB(Luria-Bertani) broth(Sigma-Aldrich)andAgar(Sigma-Aldrich))werepurchased and used without any purification. The deionized water was obtainedfromaMilliporeMilli-QUltrapureWaterSystem.Allthe chemicalswere of highpurity available and were of analytical grade.
2.2. Procurementofthebacterialstrain
Thecommercialbacterial strain utilized in this study (Pseu-domonas aeruginosa ATCC 47085) was purchased from ATCC (AmericanTypeCultureCollection,USA).Thebacterialculturewas enrichedinLBmedium(Luria-Bertani:10g/Ltryptone,5g/Lyeast extract,10g/LNaClin1Lofdistilledwater)andstockcultureswere preparedfromtheinitialbroth.Thestockcultureswerestoredat
4◦Cforshortperiodsandfreshcultureswerepreparedfromthose samplespriortothefurtheruse.
2.3. ElectrospinningofbacteriaencapsulatedPVAandPEO nanofibrouswebs
PVAandPEOnanofiberswereproducedbyusingasinglesolvent system(water),butwithdifferentpolymerconcentrations.While forPVAnanofibers,thepolymerconcentrationwas7.5%(w/v)in theelectrospinningsolution,itwas3.5%(w/v)forPEOnanofibers. Thematerialsusedforpreparationofelectrospunnanofiberswere allsterilizedbyautoclave,andtheinsideofthePlexiglasboxwhere electrospinning wascarriedout wassterilizedby UV-Clightto avoidcontamination.Beforeelectrospinningprocess,2X concen-trationsofpolymermixtures,whichweretwiceasdensedasthe regularconcentrations,werepreparedandthenequalamountsof eitherbacteria-freedistilledwaterorbacteriacontainingdistilled waterweremixedwiththesemixturestoobtain1Xelectrospinning solutions. In order to encapsulate sufficient amounts of bacte-riawithinpolymermatrices,therequiredbacterialamountwas determinedbydrycellbiomass(∼4mgofbacterialbiomassper mLofelectrospinningsolution,correspondingto∼1010cfu/mL).
The electrospinning solutions were loaded in 1mLsyringe fit-tedwith a metallic needle of 0.6mminner diameterand they werelocatedhorizontallyonasyringepump(modelKDS-101,KD Scientific,USA).Oneoftheelectrodesofhigh-voltagepower sup-ply(MatsusadaPrecision,AUSeries)wasclampedtothemetallic needle and the other one was clamped to the grounded alu-minumcollector which was covered withan aluminum foilto depositthePVAandPEOelectrospunnanofibers.The electrospin-ningparameterswereappliedas:feedrateofsolutions=1mL/h, applied voltage=10–15kV, tip-to-collector distance=10–12cm. The electrospinning apparatus was enclosed in a Plexiglas box and electrospinning wascarried out at24◦C±1 and ∼20% rel-ativehumidity.Thecollectedbacteriaencapsulatednanofibrous webswerestoredinarefrigerator(+4◦C)forquickorlonger-term use.Onlyinoneexperiment,thebacteriaencapsulatednanofibrous webswerestoredatroomtemperatureaswell.Bacteria-free (pris-tine)nanofiberswerestoredatroomtemperature.
2.4. Viabilityandstoragetests
In order to evaluate whether bacterial cells were properly encapsulatedwithin PVA and PEOnanofibers,thePseudomonas aeruginosaATCC47085cellswerestainedwithfluorescentstains (LIVE/DEADBacLightTMkit)beforemixingwiththe2X concen-trationsofpolymermixtures.Afterpreparationofelectrospinning solutions,PVAandPEOnanofiberswerecollectedonglassslides toobserveunderafluorescencemicroscope.Microscopic evalua-tionofLIVE/DEAD-stainedbacterialcellswasmadebythegeneral assessment:brightgreenfluorescenceemittingcellscorrespondto livingcellsandbrightredfluorescenceemittingcellscorrespondto deadones.PhotographsweretakenbyusingaLeicaoptical micro-scope(Leica,DMI4000B)whichhasanattachedfluorescenceunit. In addition to fluorescence microscopy, the viability of PseudomonasaeruginosaATCC 47085cellsin either electrospin-ning solutions or encapsulated within nanofibrous webs was determinedviaviablecellcounting(VCC)assay.Tofindthe encap-sulationefficiency,equivalentpiecesofthenanofibrousmaterial wasweighedwhichcontaintheencapsulatedbacterialcells. Dis-tilledwaterwasaddedtothesepiecesandtheydissolvedrapidly inwater,serial10-folddilutionsweremadeandthenthebacterial solutionswerespreadonLBagarplates.Afterovernightincubation at30◦C,thenumberofcolony-formingunits(CFU)wascounted.All testsweremadeintriplicate.
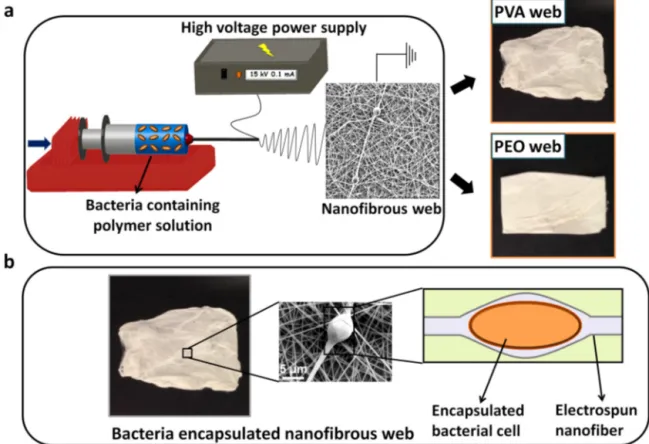
Fig.1. (a)SchematicrepresentationofelectrospinningprocessforbacteriaencapsulatedPVAandPEOwebs,andphotographsofPVAandPEOwebs,(b)representative imagesforbacteriaencapsulatedwebsincludingaSEMmicrographandaschematicrepresentationofabacterialcellinsidePVA/PEOfibers.
Forstoragetest,equivalentpieceswerepreparedforsame sam-plesandtheirviabilitieswerecheckedregularlyfordefinedtime periods.Bacteriaencapsulatednanofibrouswebsweretestedfor storabilityat4◦Cfor3monthsandat25◦Cfor10daysunderdry conditions.
2.5. Methyleneblue(MB)bioremovalexperiments
LBbrothwasutilizedasthebacterialgrowthmediumforMB bioremovalexperiments.ThepHlevelswereconstantand neu-tral(pH7.0).BacteriaencapsulatedPVAandPEOwebswereadded directlytoMBcontainingLBbrothforinitiatingbacterialgrowth. TheeffectofPVAandPEOinthegrowthmediaofdissolvedwebs onbacterialgrowthwasevaluatedbyOD600measurements.After
achievingthebacterialcellviabilitiesof each bacteria encapsu-latedPVAandPEOwebsintherangeof107–108cfu/mLper10mg,
equivalentwebsamples(withw/vratioof1mg/mL)wereprepared forinitiatingMBbioremovalexperiments.Theinitialbacterialcell viabilities of bacteria/PVA and bacteria/PEO webs were around 107–108cfu/mLand108cfu/mLper10mg,respectively.Samples
werecollectedperiodicallytoanalyzeremainingMB concentra-tionsbyaspectrophotometer,andthespecificabsorbanceofthe dyewasmeasuredat660nm.Thesampleswerecentrifugedprior tomeasurementsfor5minat10,000rpm,andthesupernatant frac-tionswereutilizedforspectrophotometricmeasurementstoavoid opticaldensity interferencefrombacterial cells.Threedifferent (10,15,25mg/L)initialMBconcentrationsweretestedfor evalu-ationoftheremovalcapabilitiesofbacteria/PVAandbacteria/PEO webs.Thesampleswereincubatedfor48hat125rpmand30◦C. Freebacteriaandbacteria-freewebsamplesweretestedfortheir MBremovalcapabilitiesaswell,aspositiveandnegativecontrols, respectively.Thebacterialcellviabilitiesofinitialinoculaforfree viablebacterialcellswereadjustedaround108cfu/mLto
compen-satetheinitialcellviabilitiesoffreeandencapsulatedbacteria.In
ordertoevaluatetheroleofdeadcellsinMBremoval,deadcells werealsotestedforMBremovalatthesameconditions.Bacterial cellviabilitieswereadjusted as∼1010 cfu/mL,correspondingto
thetotalviablebacterialnumberwithinelectrospinningsolutions beforestartingtheprocess,beforekillingbacterialcellsat70◦Cfor 3h.Alltestsweredoneintriplicate.
Theremovalcapacities(Qeq)offreebacteriacells,andbacteria
encapsulatedwebswerecalculatedbyEq.(1):
Qeq(mg/g)= (C0−−−Cf).V/M (1)
C0istheinitialMBconcentration(mg/L),Cf isthefinalMB
con-centration(mg/L),Visthesolutionvolume(L)andMisthetotal bacterialcellbiomass(g)atequilibrium[30].
2.6. Scanningelectronmicroscopy(SEM)
Millimeter-lengthPVAandPEOwebswerepreparedforSEM analysis toevaluatemorphologies of bacteria-free and bacteria encapsulatedwebs.Sampleswerecoatedwith5nmAu-Pdprior toSEMimaging(Quanta200FEGSEM,FEIInstruments,USA).The averagefiberdiameter(AFD)wasdeterminedfromtheSEMimages, andaround100fiberswereanalyzed.
2.7. Reactionkineticsstudies
TheorderofreactionsforMBremovalprocesswasevaluated upontheR2valuesofzero,first,secondandthirdorderplotsof
248 O.F.Sariogluetal./ColloidsandSurfacesB:Biointerfaces152(2017)245–251
Fig.2.SEMmicrographsof(a)pristinePVA(b)pristinePEO(c)bacteria/PVA(d)bacteria/PEOwebsandfluorescencemicroscopyimagesof(e)bacteria/PVAand(f)bacteria/PEO webs.
3. Resultsanddiscussion
3.1. Encapsulationofbacteriawithinnanofibrouswebsand evaluationofbacterialcellviability
Theelectrospinningprocessforbacteriaencapsulated nanofi-brouswebsissummarizedinFig.1schematically. Althoughthe appliedvoltageforelectrospinningprocessishighlydetrimental forbacterialcells,itwasneededtoproducePVAandPEOnanofibers atthesepolymer concentrations.Therefore, in ordertoachieve thebacterialcellviabilitiesindesiredamountsfortheelectrospun nanofibers,bacterialamountsintheelectrospinningsolutionswere highlycondensed,sothatevenafterelectrospinningandcell viabil-itylosses,thereweresufficientamountsofviablebacteriawithin electrospunnanofibers.Theencapsulationprotocolisnotanovel practiceandthereareseveralpapersrelatedwithencapsulation ofbacterialcellswithinelectrospunfibrouswebsfor bioremedi-ation[31–33].Asmentionedinthesepapers,thesurvivingcells cansuccessfullypreservetheirremovalcapabilitiesandenzymatic activitiesagainsttargetcontaminants.Inaddition,the experimen-talmethodusedinthisstudywasutilizedafterseveraloptimization steps(the appliedvoltage was determined after optimization), whichwastheleastharmfulprotocolforbacterialcellsamong
dif-ferentexperimentalconditions.Weagreethattheprocessisstill detrimentalforbacteria,yetconsiderable(andsufficientfor start-inganewculture)amountsoflivingcellscouldbeobtainedafter theprocess.Someadditionalpreservationstepscanalsobe uti-lized,forinstanceaddingglyceroltotheelectrospinningsolution toincreasethebacterialcellviabilitiesduringelectrospinning pro-cess[34],yetsinceourstudyisaproofofconceptstudy,wedidnot includethem.Furthermore,useofbiocompatibleand biodegrad-ablematerials(e.g.polyvinylalcohol,polyethyleneoxide,alginate) forencapsulationcanenhance thesurvivabilityof encapsulated cellsduringstorage.Themorphologiesofbacteria-freeand bacte-riaencapsulatedelectrospunPVAandPEOnanofibrouswebswere evaluatedbySEMimaging.TheaveragediametersofpristinePVA andPEOnanofibersweremeasuredas420±35and230±20nm, respectively.Whilebead-freenanofiberswereobtainedforpristine PVAnanofibrousweb(Fig.2a),beadedstructureswereobtained forpristinePEOnanofibrouswebat3.5%(w/v)polymer concen-tration(Fig.2b).Itwasfoundthatbacterialcellsweresuccessfully encapsulatedwithinPVAandPEOnanofibrouswebs.Encapsulation ofbacterialcellscausedlocalwideningoffibersincertainregions andball-likestructureswereobservedintheseareas(Fig.2cand d).Inaddition,itwasnoticedthattheball-likestructuresdueto bacterialencapsulationarequitedifferent(biggerandthicker)and
Table1
Removalcapacitiesoffree-bacteria,bacteria/PVAwebandbacteria/PEOweb sam-plesatequilibriumattheendoftheremovalprocess.T=30◦C,agitationrate: 125rpm,incubationtime:48h(n=3,resultsshowmeans±S.E.M).
Samplename Initialconcentration(C0) Qeq(mg/g)
Free-bacteria 10mg/L 25.04±0.58 15mg/L 66.75±6.59 25mg/L 74.02±8.61 Bacteria/PVAweb 10mg/L 27.79±3.18 15mg/L 45±1.38 25mg/L 89.22±1.17 Bacteria/PEOwe 10mg/L 23.57±3.24 15mg/L 60.04±1.46 25mg/L 89.47±3.94
canbeeasilydifferentiatedfromtheordinarybeadsofpristinePEO
nanofibers(Fig.2d).Fluorescencemicroscopyimageshaveshown
that,thecellviabilitieswerepreservedfortheencapsulated bacte-riainthePVAandPEOnanofibermatrices(Fig.2eandf).
The bacterial cell viabilities were also checked by applying VCCassayonequivalentsamplesofbacteria/PVAandbacteria/PEO webs.Asmentionedpreviously,beforeusinginMBremoval exper-iments, thecell viabilities were determined for each 10mg of bacteriaencapsulatedwebsasaround107–108cfu/mLfor
bacte-ria/PVAand108cfu/mLforbacteria/PEOwebs.Bacteria/PEOweb
samples had always higher cell viabilities, possibly due tothe higherw/vratioofbacterialcellbiomassafterdehydrationfor bac-teria/PEOwebs.Afterensuringthatthewebsampleshavesufficient amountsofviablebacterialcells,biodegradationexperimentswere initiatedwithequivalentsofthosewebsamples.
3.2. MBdyeremovalcapabilitiesofbacteria/PVAand bacteria/PEOwebs
Electrospun bacteria/PVAand bacteria/PEO webs arereadily water-solublebiocompositeswhichcouldbehandycarriermatrix forbacterialstorageandcanbealternativetolyophilizedbacteria forenvironmentalremediationapproachesinwater.Theeffectof PVAandPEOpolymericsolutionsonbacterialgrowthwas evalu-atedandnoapparentdifferenceswerefoundforbacteriawhich weregrowninpolymericsolutions(Fig.S1),hencethesewebscan beusedsafelyforstartingbacterialinocula.Here,MBremoval capa-bilitiesofbacteria/PVAandbacteria/PEOwebsweretestedatthree differentinitialMBconcentrations(10,15,25mg/L).Whileboth webshaveshownlowerremovalyieldsat10mg/LofinitialMB (52.5%forbacteria/PVAweband44.4%forbacteria/PEOweb),their removalyieldsincreasedat15mg/L(57%forbacteria/PVAweband 76%forbacteria/PEOweb)and25mg/L(68%forbacteria/PVAweb and69%forbacteria/PEOweb)ofinitialMB,suggestingtheremoval processisbiologicalratherthanadsorptionbased,andmightbe enhancedbygeneticswitchingatadefinedconcentrationrange (Fig.3a).EventhoughPVAandPEOnanofibrouswebswerequickly dissolvedinMBaqueoussolution,averynegligibledecreasewas observedinMBconcentrationwiththeadditionofpristinePVA andPEOnanofibers,implyingtheremovalperformancesof bac-teria/PVA and bacteria/PEO webs were primarilybased onthe bacterialexistence(Fig.3b).Inaddition,deadbacterialcellshave shownaverynegligibledecreaseintheinitialMBconcentration after48hincubation,suggesting MBdyewasprimarily remedi-atedbyviablebacterialcells(Fig.S2).Removalcapacities(Qeq)of
free-bacteriacellsandbacteriaencapsulatedwebswerecalculated foreachconcentrationandarepresentedinTable1.TheQeqvalues
offree-bacteria,bacteria/PVAwebandbacteria/PEOwebsamples aresimilarat10mg/L.At15mg/LofinitialMBconcentration, free-bacteriasamplehasahigherQeqvaluethanbacteriaencapsulated
webs,andbacteria/PEOweb hasahigherQeq valuethan
bacte-ria/PVAweb.At25mg/LofinitialMBconcentration,theQeqvalues
ofbacteria/PVAandbacteria/PEOwebsareveryclosetoeachother andbothofthemarehigherthanthefree-bacteriasample.Since bacteria/PEOwebsampleshadhigheramountsofviablebacteriafor theinitialinoculum,theirremovalperformanceswerehigherthan thatofbacteria/PVAwebsamplesingeneral,suggestingthe bacte-riaencapsulatedwebsamplescanbeimprovedformoreefficient MBremovalbyincreasingtheencapsulatedbacterialcell viabili-ties.Inaddition,byusingamorecapablebacterialstrainforMB removal,higherremovalperformanceseveninshortertime peri-odscanbeobtained,aspresentedpreviously[1,35].Notaregular trendwasobservedwhencomparingtheremovalperformancesof free-bacterialcellsandbacteriaencapsulatedwebsamples.While thehighest MBremovalwasobserved byfree-bacterial cellsat 15mg/L,thehighestMBremovalwasobservedbybacteria/PEO websampleat25mg/L,andthehighestMBremovalwasobserved bybacteria/PVAwebsample at10mg/L.Theseresultsmight be occurredduetothedifferencesinencapsulationefficiencyfor dif-ferentbacteriaencapsulatedwebsamples,leadingdifferencesin initialinoculaandhence maximalgrowthofthebacterialcells. Sinceencapsulationefficiencycanbeinfluencedbatchtobatchdue toslightenvironmentalchanges(e.g.humidity),somefluctuations inbacterialnumberswereobservedforequivalentsamples.
3.3. Evaluationoforderofreactions
TheR2valuesofdifferentorderplotsforMBremovalarelisted
inTableS1.Whilebacteria/PVAwebhasshownthehighest cor-relationwiththezeroordermodel(R2=0.9797),free-bacteriaand
bacteria/PEOwebsampleshaveshownthehighestcorrelationwith thefirstordermodel(R2valuesof0.9912and0.943,respectively).
Theseresultsconformwiththeresultsfromtheliterature,since enzyme-catalyzedreactionsoftenfallunderthezeroordermodel
[36]andfirstorderreactionscanfittotheenzyme-drivenreactions forbiologicalremovalofwatercontaminants[21].
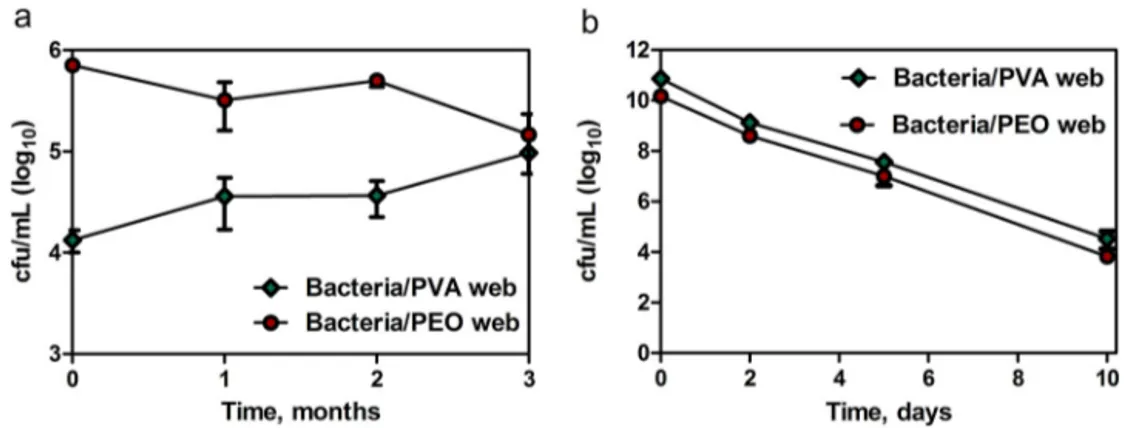
3.4. Storabilityandapplicabilityofbacteriaencapsulatedwebs Thebacteriaencapsulatedwebsamplesweretestedfor storabil-ityatdifferenttimeperiodsviaVCCassay,intermsofcellviability preservation.Different levelsof cellviabilitieswereachievedin two different experiments(at 4◦C and 25◦C)and bacteria/PVA websamplehadhigherinitialcellviabilitythanbacteria/PEOweb sampleforthestoragetestat25◦C(Fig.4b),unlikefromthe pre-viousexperiments,probablyduetoaninfluentialbatchtobatch variation.Nevertheless,thesedifferencesdidnothavesignificant impactsontheassessmentofstoragetest,sinceeachsamplehas beenevaluatedindividuallyandthecomparisonsweremadeon theirinitialtofinalcellviabilityratios.Inaddition,althoughthere aresomevariationsinthecellviabilitymeasurementsofbacteria encapsulatedweb samplesforthestoragetestat4◦C,theseare restrictedincertainlevels,henceitwassupposedthatthepartial dissimilaritiesinthecellviabilitynumbersforequivalentsamples mightbethereasonforthesevariations.Itwasfoundthat,while bacteriaencapsulatedwebsamplescanbesafelystoredat4◦Cfor monthswithoutsignificantlossesintheinitialcellviabilities,the cellviabilitiesrapidlydecreaseat25◦Canddonotallowlong-term storageatthistemperature(Fig.4,TableS2).Therefore,itwas con-cludedthat,thebacteriaencapsulatedwebsamplescanbestored safelyforlongtimeperiods,yetitneedscoolertemperaturesfor cellviabilitypreservation.
Inbrief,thisstudyfocusesondesign anddevelopmentofan alternativesystemwithdistinctfeatures,ratherthantheremoval efficiencyforMBremediation.Totheauthors’knowledge,thisis thefirststudywhichpresentsremediationofMBbybacteria encap-sulatedelectrospunfibrouswebs.Thebacteriaencapsulatedwebs
250 O.F.Sariogluetal./ColloidsandSurfacesB:Biointerfaces152(2017)245–251
Fig.3. (a)MBremovalprofilesoffree-bacteria,bacteria/PVAwebandbacteria/PEOwebsamplesatinitialconcentrationsof10,15and25mg/L.(b)Concentrationvs.time graphoffree-bacteria,bacteria/PVAweb,bacteria/PEOweb,pristinePVAwebandpristinePEOwebsamplesat15mg/LofinitialMB(n=3,graphsshowmeans±S.E.M).
Fig.4.Viablecellcounting(VCC)resultsofbacteria/PVAandbacteria/PEOwebsamplesforstorageat(a)4◦Cfor3monthsand(b)25◦Cfor10days(n=3,graphsshow
means±S.E.M).
havelowerspaceandweightcomparetofree-bacteriain liquid media,whichprovideseaseofapplicationandlower transporta-tioncosts,asinlyophilizedbacteria.Inaddition,thesewebscanbe storedatcoolertemperatureswithoutsignificantlossesinthecell viability.Byoptimizationofenvironmentalparametersandusinga morecapablebacterialstrainintermsofremovalefficiencyagainst MBdye,moreefficientbiocompositescanbeproducedfor remedia-tionofMB.Inthissense,thefindingsherearepromisingforfurther developmentsinthisfield.
4. Conclusions
Inthisstudy,wehavedevelopedfunctionalbiocomposite mate-rialsthat wereproduced byencapsulationofa MBremediating PseudomonasaeruginosastrainwithinelectrospunPVAand PEO nanofibrouswebs.Thebacterialcellviabilitieswerecheckedby viable cell counting (VCC) assay and fluorescence microscopy imaging.Sufficientamountsofviablebacterialcellscouldbe encap-sulatedwithinelectrospunPVAandPEOnanofibrouswebs,and thesewebsweretestedforMBremovalinwater.TheresultsofMB removalexperimentsrevealedthatMBremovalcapabilitiesof bac-teriaencapsulatedwebswerebasedonthebacterialpresence,and similarremovalperformanceswereobservedforfree-bacteria.It wasinferredthatMBremovalwasachievedbybiologicalremoval ratherthanadsorption,andtheremovalperformancescanbe opti-mized by increasing the initialcell viability numbers or using amore capablebacterialstrain.Inaddition, storagetestresults showedthatbacteriaencapsulatedwebscanbestoredsafelyfor longtimeperiodsat4◦C,whilepreservingtheinitialcell viabil-itynumbers.Thistypeofstoragecanbealternativetolyophilized bacteriaandbringsomeadvantagesoverstorageinculturemedia;
suchasthewebsamplesdonotneedanyminimalgrowthmedium anditrequiresverysmallspacesforstorage.Inconclusion, bacte-riaencapsulatedelectrospunnanofibrouswebscanbeeffectively usedforremediationofMBinwaterwithstorableandimprovable properties.
Acknowledgements
The Scientific and Technological Research Councilof Turkey (TUBITAK, project #114Y264) is acknowledged for fundingthe research.Dr.UyaracknowledgesTheTurkishAcademyofSciences - Outstanding Young Scientists Award Program (TUBA-GEBIP) for partialfundingof theresearch. A.Celebioglu acknowledges TUBITAKproject#113Y348forapostdoctoralfellowship.O.F. Sar-iogluacknowledgesTUBITAK BIDEB(2211-C)for NationalPh.D. Scholarship.
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound,in theonlineversion,athttp://dx.doi.org/10.1016/j.colsurfb.2017.01. 034.
References
[1]N.O.San,A.Celebioglu,Y.Tümtas¸,T.Uyar,T.Tekinay,Reusablebacteria immobilizedelectrospunnanofibrouswebsfordecolorizationofmethylene bluedyeinwastewatertreatment,RSCAdv.4(2014)32249–32255.
[2]N.S.Maurya,A.K.Mittal,P.Cornel,E.Rother,Biosorptionofdyesusingdead macrofungi:effectofdyestructure:ionicstrengthandpH,Bioresour. Technol.97(2006)512–521.
[3]A.Saeed,M.Iqbal,S.I.Zafar,ImmobilizationofTrichodermaviridefor enhancedmethylenebluebiosorption:batchandcolumnstudies,J.Hazard. Mater.168(2009)406–415.
[4]K.Vijayaraghavan,S.W.Won,J.Mao,Y.S.Yun,Chemicalmodificationof Corynebacteriumglutamicumtoimprovemethylenebluebiosorption,Chem. Eng.J.145(2008)1–6.
[5]V.J.P.Vilar,C.M.S.Botelho,R.A.R.Boaventura,Methyleneblueadsorptionby algalbiomassbasedmaterials:biosorbentscharacterizationandprocess behavior,J.Hazard.Mater.147(2007)120–132.
[6]X.Wang,X.Chen,K.Yoon,D.Fang,B.S.Hsiao,B.Chu,Highfluxfiltration mediumbasedonnanofibroussubstratewithhydrophilicnanocomposite coating,Environ.Sci.Technol.39(2005)7684–7691.
[7]N.Zaghbani,A.Hafiane,M.Dhahbi,Separationofmethylenebluefrom aqueoussolutionbymicellarenhancedultrafiltration,Sep.Purif.Technol.55 (2007)117–124.
[8]R.Wang,J.Guo,D.Chen,Y.E.Miao,J.Pan,W.W.Tjiu,T.Liu,Tubebrushlike ZnO/SiO2hybridtoconstructaflexiblemembranewithenhanced
photocatalyticpropertiesandrecyclingability,J.Mater.Chem.21(2011) 19375–19380.
[9]Y.Zhan,H.Li,Y.Chen,Copperhydroxyphosphateascatalystforthewet hydrogenperoxideoxidationofazodyes,J.Hazard.Mater.180(2010) 481–485.
[10]S.K.Nataraj,K.M.Hosamani,T.M.Aminabhavi,Distillerywastewater treatmentbythemembrane-basednanofiltrationandreverseosmosis processes,WaterRes.40(2006)2349–2356.
[11]E.Rosales,M.Pazos,M.A.Sanromán,Comparativeefficienciesofthe decolourisationofleatherdyesbyenzymaticandelectrochemicaltreatments, Desalination278(2011)312–317.
[12]A.Malik,Metalbioremediationthroughgrowingcells,Environ.Int.30(2004) 261–278.
[13]M.T.Madigan,J.M.Martinko,BrockBiologyofMicroorganisms,eleventhed., PrenticeHall,NewJersey,2006.
[14]A.K.Karamalidis,A.C.Evangelou,E.Karabika,A.I.Koukkou,C.Drainas,E.A. Voudrias,Laboratoryscalebioremediationofpetroleum-contaminatedsoilby indigenousmicroorganismsandaddedPseudomonasaeruginosastrainSpet, Bioresour.Technol.101(2010)6545–6552.
[15]Y.Wang,J.Song,W.Zhao,X.He,J.Chen,M.Xiao,Insitudegradationofphenol andpromotionofplantgrowthincontaminatedenvironmentsbyasingle Pseudomonasaeruginosastrain,J.Hazard.Mater.192(2011)354–360.
[16]R.Pasumarthi,S.Chandrasekaran,S.Mutnuri,Biodegradationofcrudeoilby PseudomonasaeruginosaandEscherichiafergusoniiisolatedfromtheGoan coast,Mar.Pollut.Bull.76(2013)276–282.
[17]E.Eroglu,V.Agarwal,M.Bradshaw,X.Chen,S.M.Smith,C.L.Raston,K.S.Iyera, Nitrateremovalfromliquideffluentsusingmicroalgaeimmobilizedon chitosannanofibermats,GreenChem.14(2012)2682–2685.
[18]L.Hall-Stoodley,J.W.Costerton,P.Stoodley,Bacterialbiofilms:fromthe Naturalenvironmenttoinfectiousdiseases,Nat.Rev.Microbiol.2(2004) 95–108.
[19]O.F.Sarioglu,O.Yasa,A.Celebioglu,T.Uyar,T.Tekinay,Efficientammonium removalfromaquaticenvironmentsbyAcinetobactercalcoaceticusSTB1 immobilizedonanelectrospuncelluloseacetatenanofibrousweb,Green Chem.15(2013)2566–2572.
[20]N.O.San-Keskin,A.Celebioglu,T.Uyar,T.Tekinay,Microalgaeimmobilizedby nanofibrouswebforremovalofreactivedyesfromwastewater,Ind.Eng. Chem.Res.54(2015)5802–5809.
[21]N.O.San-Keskin,A.Celebioglu,O.F.Sarioglu,A.D.Ozkan,T.Uyar,T.Tekinay, Removalofareactivedyeandhexavalentchromiumbyareusablebacteria attachedelectrospunnanofibrousweb,RSCAdv.5(2015)86867–86874.
[22]O.F.Sarioglu,A.Celebioglu,T.Tekinay,T.Uyar,Evaluationofcontacttimeand fibermorphologyonbacterialimmobilizationfordevelopmentofnovel surfactantdegradingnanofibrouswebs,RSCAdv.5(2015)102750–102758.
[23]W.Salalha,J.Kuhn,Y.Dror,E.Zussman,Encapsulationofbacteriaandviruses inelectrospunnanofibres,Nanotechnology17(2006)4675–4681.
[24]W.Y.Fung,K.H.Yuen,M.T.Liong,Agrowaste-basednanofibersasaprobiotic encapsulant:fabricationandcharacterization,J.Agric.FoodChem.59(2011) 8140–8147.
[25]Y.Liu,M.H.Rafailovich,R.Malal,D.Cohn,D.Chidambaram,Engineeringof bio-hybridmaterialsbyelectrospinningpolymer-microbefibers,Proc.Natl. Acad.Sci.U.S.A.106(2009)14201–14206.
[26]S.Klein,R.Avrahami,E.Zussman,M.Beliavski,S.Tarre,M.Green, EncapsulationofPseudomonassp:ADPcellsinelectrospunmicrotubesfor atrazinebioremediation,J.Ind.Microbiol.Biotechnol.39(2012)1605–1613.
[27]H.W.Tong,B.R.Mutlu,L.P.Wackett,A.Aksan,Manufacturingofbioreactive nanofibersforbioremediation,Biotechnol.Bioeng.111(2014)1483–1493.
[28]N.Jiang,G.L.Ying,S.Y.Liu,L.Shen,J.Hu,L.J.Dai,X.Y.Yang,G.Tian,B.L.Su, Aminoacid-basedbiohybridsfornano-shellizationofindividualdesulfurizing bacteria,Chem.Commun.50(2014)15407–15410.
[29]X.Y.Yang,G.Tian,N.Jiang,B.L.Su,Immobilizationtechnology:asustainable solutionforbiofuelcelldesign,EnergyEnviron.Sci.5(2012)5540–5563.
[30]C.J.Buchko,L.C.Chen,Y.Shen,D.C.Martin,Processingandmicrostructural characterizationofporousbiocompatibleproteinpolymerthinfilms,Polymer 40(1999)7397–7407.
[31]H.W.Tong,B.R.Mutlu,L.P.Wackett,A.Aksan,Silica/PVAbiocatalytic nanofibers,Mater.Lett.111(2013)234–237.
[32]S.Klein,J.Kuhn,R.Avrahami,S.Tarre,M.Beliavski,M.Green,E.Zussman, Encapsulationofbacterialcellsinelectrospunmicrotubes,
Biomacromolecules10(2009)1751–1756.
[33]S.Klein,R.Avrahami,E.Zussman,M.Beliavski,S.Tarre,M.Green, EncapsulationofPseudomonassp.ADPcellsinelectrospunmicrotubesfor atrazinebioremediation,J.Ind.Microbiol.Biotechnol.39(2012)1605–1613.
[34]W.Salalha,J.Kuhn,Y.Dror,E.Zussman,Encapsulationofbacteriaandviruses inelectrospunnanofibres,Nanotechnology17(2006)4675–4681.
[35]N.A.El-Sersy,BioremediationofmethylenebluebyBacillusthuringiensis4G1: applicationofstatisticaldesignsandsurfaceplotsforoptimization, Biotechnology6(2007)34–39.
[36]I.Tinoco,K.Sauer,J.C.Wang,PhysicalChemistry–PrinciplesandApplications inBiologicalSciences,thirded.,PrenticeHall,NewJersey,1996.