ANKARA ÜNİVERSİTESİ ZİRAAT FAKÜLTESİ
Genetic Analysis of Grapevine Cultivars from the Eastern
Mediterranean Region of Turkey, Based on SSR Markers
Serpil Gök TANGOLAR1 Semra SOYDAM2 Melike BAKIR3 Eda KARAAĞAÇ3 Semih TANGOLAR4 Ali ERGÜL3
Geliş Tarihi: 24.11.2008 Kabul Tarihi: 30.03.2009
Abstract : Turkey is one of the centres for grapevine diversity with many local grapevine cultivars with potentially desirable characteristics are still widely cultivated throughout several regions in the country. One of such regions is the Eastern Mediterranean region of the country. To characterize the grapevine germplasm of this region, we performed a genetic analysis of 59 grapevine cultivars using 14 simple sequence repeat (SSR) loci. The genetic relationships of the cultivars which did not appear to be linked to their ecogeographical distributions within the region were determined. Among the cultivars analyzed in this study, several synonymous and homonymous cultivars were identified, suggesting that some of the grapevine cultivars with different names might be genetically identical while some cultivars with the same names might be genetically different. This study represents a first step toward the genetic characterization of the grapevine genotypes of this region and will help promote more effective germplasm management and breeding programs.
Key Words: Vitis vinifera L., SSR, Eastern Mediterranean, synonym, homonym.
Türkiye Doğu Akdeniz Bölgesi Üzüm Çeşitlerinin SSR Markörleri ile
Genetik Analizi
Öz: Türkiye, asma genetik kaynaklarının önemli merkezlerinden birisidir. Ülkenin bazı bölgelerinde hala yaygın şekilde yetiştiriciliği yapılan birçok yöresel üzüm çeşidi bulunmaktadır. Bu bölgelerden biri Doğu Akdeniz Bölgesidir. Bu çalışmada, bölgenin asma gen kaynaklarının tanımlanması amacı ile 14 SSR primeri kullanılarak 59 üzüm çeşidinin genetik analizleri yapılmıştır. Çalışma sonucunda çeşitler arasındaki genetik ilişkilerin bölge içindeki ekocoğrafik dağılımları ile bağlantılı olmadığı belirlenmiştir. Çalışmada analiz edilen çeşitler arasında farklı isimlerle anıldığı halde aynı genotip (sinonim) veya aynı isimli ancak farklı genotip özelliği (homonim) gösterenler saptanmıştır. Bölgenin asma genotipleri ile ilgili ilk genetik analiz sonuçlarının sunulduğu bu çalışma, gen kaynaklarının daha etkin kullanımını ve ıslah programlarını destekleyecektir.
Anahtar Kelimeler: Vitis vinifera L., SSR, Doğu Akdeniz, sinonim, homonym.
Introduction
Turkey is a center of diversity for grapevines (Vitis vinifera L.) and possesses a rich grapevine germplasm (Arroya-Garcia et al. 2006, Ergül et al. 2006). The Mediterranean region of Turkey with suitable climatic conditions possesses the one of the largest areas devoted to grapevine production in the country, second only to the Aegean region. Whereas early maturing cultivars are common in the low-altitude regions (i.e. the region spanning central Adana and İçel, two provinces that are separated by the Tarsus district), late-maturing table grape and wine cultivars are more prevalent at higher altitudes (e.g., the Saimbeyli, Feke and Pozanti districts) of this region.
As expected, viticulture is also a valuable economic activity in most of the country, except for the high altitude regions of East Anatolia and the Eastern Black Sea where unsuitable climatic conditions (e.g. excessive rainfall) largely restrict grapevine cultivation. Grapevine cultivation and wine culture have long been important elements of life in the Eastern Mediterranean region (Oraman 1969) with many local cultivars are still widely cultivated. However, given the relatively large numbers of such cultivars grown in the region, problems related to cultivar identity have recently begun to appear within this germplasm, such that certain cultivars are grown under different names, 1Ministry of Agriculture and Rural Affairs, Plant Protection Research Institute- Adana
2Ankara Univ,, Fac. of Science, Department of Biology-Ankara 3Ankara Univ., Biotechnology Institute-Ankara
2 TARIM BİLİMLERİ DERGİSİ 2009, Cilt 15, Sayı 1
whereas other, more discrete, cultivars are cultivated under the same name. Consumer preference for certain grape cultivars has increased the possibility that local cultivars which have been widely cultivated for centuries and are well adapted to the region, would eventually disappear.
In this study, we have performed a genetic characterization of grape cultivars grown in the Eastern Mediterranean region (i.e. Adana and the surrounding districts Gülnar, İçel and Tarsus), as well as other Mediterranean cultivars that are well adapted to this region. Specifically, we analyzed 14 simple sequence repeats (SSR) loci from 59 grapevine cultivars and examined allelic data to determine the genetic relationships among these cultivars. Our findings will assist with the conservation of these cultivars and help alleviate problems related to cultivar identity.
Materials and Methods
Plant materials: Plant samples were collected
from 15-year-old grapevines maintained at Çukurova
University Faculty of Agriculture Collection Vineyard in Adana, as well as from privately owned vineyards (these plants were then included in the collection
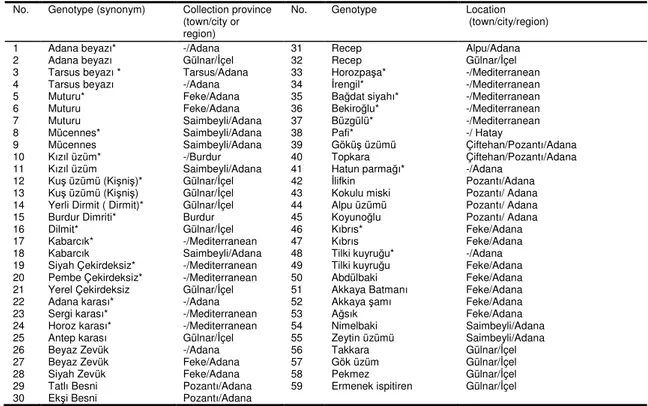
vineyard in 2008). Grapevine cultivars used in this study and their collection sites are shown in Table 1.
DNA extraction: DNA was extracted using the
procedure described by Lefort et al. (1998) and Ağaoğlu et al. (2001). Concentration and purity of the extracted DNA were analyzed using a NanoDrop® ND-1000 spectrophotometer.
Genetic analysis of simple sequence repeats:
We analyzed 14 SSR markers, of which 6 belonged to the so-called “core set” (i.e., VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, and VrZAG79) of markers that are recommended for the direct comparison of results from different laboratories (This et al. 2004). PCR amplifications were performed as described by Şelli et al. (2007). PCR products were diluted in sample loading solution (SLS) and standards from the Genomelab DNA Standard Kit-400 were included. The amplified fragments were analyzed at least twice using a CEQ 8800XL capillary DNA analysis system (Beckman Coulter, Fullerton, CA) to ensure reproducibility. Allele sizes were determined for each SSR locus using the Beckman CEQ fragment analysis software. In each run, Cabernet Sauvignon and Merlot were included as reference cultivars.
Table 1. Origin of grape cultivars used in this study
No. Genotype (synonym) Collection province (town/city or region)
No. Genotype Location (town/city/region) 1 Adana beyazı* -/Adana 31 Recep Alpu/Adana 2 Adana beyazı Gülnar/İçel 32 Recep Gülnar/İçel 3 Tarsus beyazı * Tarsus/Adana 33 Horozpaşa* -/Mediterranean 4 Tarsus beyazı -/Adana 34 İrengil* -/Mediterranean 5 Muturu* Feke/Adana 35 Bağdat siyahı* -/Mediterranean 6 Muturu Feke/Adana 36 Bekiroğlu* -/Mediterranean 7 Muturu Saimbeyli/Adana 37 Büzgülü* -/Mediterranean 8 Mücennes* Saimbeyli/Adana 38 Pafi* -/ Hatay
9 Mücennes Saimbeyli/Adana 39 Göküş üzümü Çiftehan/Pozantı/Adana 10 Kızıl üzüm* -/Burdur 40 Topkara Çiftehan/Pozantı/Adana 11 Kızıl üzüm Saimbeyli/Adana 41 Hatun parmağı* -/Adana
12 Kuş üzümü (Kişniş)* Gülnar/İçel 42 İlifkin Pozantı/Adana 13 Kuş üzümü (Kişniş) Gülnar/İçel 43 Kokulu miski Pozantı/ Adana 14 Yerli Dirmit ( Dirmit)* Gülnar/İçel 44 Alpu üzümü Pozantı/ Adana 15 Burdur Dimriti* Burdur 45 Koyunoğlu Pozantı/ Adana 16 Dilmit* Gülnar/İçel 46 Kıbrıs* Feke/Adana 17 Kabarcık* -/Mediterranean 47 Kıbrıs Feke/Adana 18 Kabarcık Saimbeyli/Adana 48 Tilki kuyruğu* -/Adana 19 Siyah Çekirdeksiz* -/Mediterranean 49 Tilki kuyruğu Feke/Adana 20 Pembe Çekirdeksiz* -/Mediterranean 50 Abdülbaki Feke/Adana 21 Yerel Çekirdeksiz Gülnar/İçel 51 Akkaya Batmanı Feke/Adana 22 Adana karası* -/Adana 52 Akkaya şamı Feke/Adana 23 Sergi karası* -/Mediterranean 53 Ağsık Feke/Adana 24 Horoz karası* -/Mediterranean 54 Nimelbaki Saimbeyli/Adana 25 Antep karası Gülnar/İçel 55 Zeytin üzümü Saimbeyli/Adana 26 Beyaz Zevük -/Adana 56 Takkara Gülnar/İçel 27 Beyaz Zevük Feke/Adana 57 Gök üzüm Gülnar/İçel 28 Siyah Zevük Feke/Adana 58 Pekmez Gülnar/İçel 29 Tatlı Besni Pozantı/Adana 59 Ermenek ispitiren Gülnar/İçel 30 Ekşi Besni Pozantı/Adana
*Plant samples were collected from Experimental Vineyard at Çukurova University, Faculty of Agriculture, Adana. -: information is not available.
The number of alleles (n), allele frequency, expected (He) and observed (Ho) heterozygosity, estimated frequency of null alleles (r), probability of identity (PI), and presence of identical genotypes were determined for each locus using IDENTITY version 1.0 software (Wagner and Sefc 1999), as described by Paetkau et al. (1995). Microsat version 1.5 (Minch et al. 1995) was used to calculate the proportion of shared alleles with the ps option [i.e., option 1-(ps)] (Bowcock et al. 1994) selected to assess genetic dissimilarity. Data were then converted to a similarity matrix, and a dendogram was constructed via the unweighted pair-group with arithmetic mean (UPGMA) method (Sneath and Sokal 1973), using the numerical Taxonomy and Multiware Analysis System (NTSYS-pc) software, version 2.0 (Rohlf1998).
Results and Discussion
SSR analysis: We analyzed 14 SSR markers
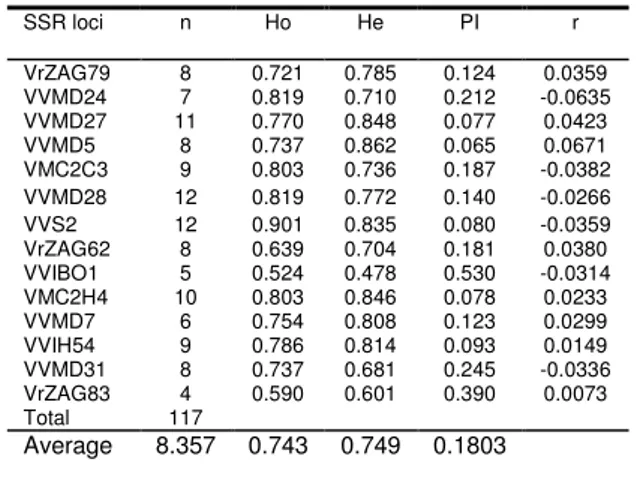
from 59 grapevine cultivars grown in Adana and neighboring regions (in addition to two reference cultivars) and detected a total of 117 alleles. The average number of alleles was 8.357, consistent with data reported for other grapevines (Dangl et al. 2001, Costantini et al. 2005) (Table 2).
The mean observed heterozygosity (Ho) and expected heterozygosity (He) were determined to be 0.743 and 0.749, respectively. The Ho values of the VVMD24, VMC2C3, VVMD28, VVS2, VVIBO1 and VVMD31 loci were high. Previous reports also found relatively high Hes in some other grape loci (Costantini et al. 2005, Santana et al. 2007). In this study, the frequency of null alleles (r) at these VrZAG79, VVMD27, VVMD5, VrZAG62, VMC2H4, VVMD7, VVIH54 and VrZAG83 loci was positive, but these low values suggest the absence of null alleles (Table 2).
The VrZAG83 locus generated the lowest number of alleles (4 alleles), whereas the VVMD28 and VVS2 loci generated the highest number of alleles (12 alleles). Similarly, previous studies have reported that VVS2 (Fatahi et al. 2003, Núñez et al. 2004, Şelli et al. 2007) and VVMD28 generate the highest number of alleles (Vouillamoz et al. 2006, Şelli et al. 2007). Other studies, including ours, have also reported that VrZAG83 generates the lowest number of alleles (Sefc et al. 2000, Snoussi et al. 2004) (Table 2).
The most informative loci, with regards to the probability of identity (PI), were VVMD28 and VVS2, which each generated 12 alleles (PI: 0.132 and 0.084, respectively). The least informative locus was VrZAG83, which generated four alleles (PI: 0.389)
(Table 2). Allele sizes (bp) of 59 cultivars from 14 SSR loci are shown in Table 3.
Identification of synonyms and homonyms:
Many researchers have used SSR markers to identify synonyms and homonyms of grapevine genotypes (Ibáñez et al. 2003, Martín et al. 2003, This et al. 2004, Vouillamoz et al. 2006, Şelli et al. 2007). Among the grape cultivars analyzed in this study, three cases of identity were found [Muturu (5), Muturu (6) and Muturu (7); Kuş üzümü (Kişniş) (12) and Kuş üzümü (Kişniş) (13); and Kabarcık (17) and Kabarcık (18)] and five cases of synonyms were identified [Adana beyazı (2) and Recep (31); Kuş üzümü (Kişniş) (12 and 13), Dilmit (16) and Topkara (40); Ekşi besni (30) and Tilki kuyruğu (48); Göküş üzümü (39) and Koyunoğlu (45); and Abdülbaki (50) and Nimelbaki (54)].
Our results also indicated that a number of cultivars known by the same names were genetically different, suggesting that they were homonyms. These genotypes are Adana beyazı (1) and Adana beyazı (2); Tarsus beyazı (3) and Tarsus beyazı (4); Mücennes (8) and Mücennes (9); Kızıl üzüm(10) and Kızıl üzüm (11); Beyaz Zevük (26) and Beyaz Zevük (27); Recep (31) and Recep (32); Kıbrıs (46) and Kıbrıs (47); and Tilki kuyruğu (48) and Tilki kuyruğu (49).
SSR-based relationship and ecogeographical
distributions: When the genetic similarity of the genotypes was analyzed, the highest similarity ratio (not including the identical and synonym cultivars) was 96.4% between 10 genotypes; however, the rest of the genotypes revealed a similarity index below 92.9%.
The dendrogram shown in Figure 1 reveals that the reference cultivars used in this study are distantly related to the Turkish cultivars studied here. The distribution of cultivars within the dendrogram appears to be independent of their ecogeographical distributions. Among cultivars with similar names,
some clustered together (e.g., Muturu, Mücennes, Kuş
üzümü, Kabarcık, and Kıbrıs), whereas others [e.g.,
Adana beyazı, Tarsus beyazı, Kızıl üzüm, Dilmit, Çekirdeksiz, Zevük, Besni, Recep, and Tilki kuyruğu] were more distantly related.
We identified two genotypes of Tarsus beyazı (an early-maturing grape cultivar with a white berry color), which is the most common early grape cultivar native to Adana. These data suggest that genetic variation exists within this cultivar. In contrast, other early-maturing grape cultivars found in the region [Tarsus beyazı (3) and Adana karası (22)] displayed a high similarity index (almost 80%), suggesting that they might have originated from the same genetic background.
4 TARIM BİLİMLERİ DERGİSİ 2009, Cilt 15, Sayı 1 Table 2. SSR loci and genetic characteristics of grape
cultivars. SSR loci n Ho He PI r VrZAG79 8 0.721 0.785 0.124 0.0359 VVMD24 7 0.819 0.710 0.212 -0.0635 VVMD27 11 0.770 0.848 0.077 0.0423 VVMD5 8 0.737 0.862 0.065 0.0671 VMC2C3 9 0.803 0.736 0.187 -0.0382 VVMD28 12 0.819 0.772 0.140 -0.0266 VVS2 12 0.901 0.835 0.080 -0.0359 VrZAG62 8 0.639 0.704 0.181 0.0380 VVIBO1 5 0.524 0.478 0.530 -0.0314 VMC2H4 10 0.803 0.846 0.078 0.0233 VVMD7 6 0.754 0.808 0.123 0.0299 VVIH54 9 0.786 0.814 0.093 0.0149 VVMD31 8 0.737 0.681 0.245 -0.0336 VrZAG83 4 0.590 0.601 0.390 0.0073 Total 117 Average 8.357 0.743 0.749 0.1803
Abbreviations: N, number of alleles; Ho, observed heterozygosity; He, expected heterozygosity; PI, probability; r, null allele frequencies.
Two genotypes of Adana beyazı (a cultivar with a white berry color), which is also grown extensively in the region, revealed a low similarity index (approximately 35.7%). On the other hand, a high degree of similarity was observed between Adana beyazı (2) and Recep (a cultivar with a white berry color) genotypes 31 and 32 (100% and 64.3%, respectively), suggesting that Adana beyazı (2) was improperly named.
Tilki kuyruğu (48) is thought to be distantly related to Tilki kuyruğu (49). However, our data suggest that Tilki kuyruğu (48) is a synonym of Ekşi besni (30) (a cultivar with a white berry color), suggesting that this cultivar is also improperly named and should be reclassified as Ekşi besni.
Genotypes 16 (Dilmit) and 40 (Topkara) were found to be synonyms of Kuş üzümü (Kişniş) (12 and 13), suggesting that the Dilmit and Topkara cultivars might be in fact Kuş üzümü.
Figure 1 . Genetic relationships among cultivars based on variation detected at 14 SSR loci.
Genetic Similarity 0.25 0.44 0.62 0.81 1.00 1 15 14 20 10 3 22 5 6 7 52 12 13 16 40 51 34 37 25 56 4 11 17 18 42 29 57 2 31 32 39 45 59 49 50 54 26 30 48 27 38 8 53 9 58 24 44 28 41 43 23 55 35 46 47 19 21 33 36 CS M
T ab le 3 . A lle le s iz es ( bp ) fo r 59 c ul tiv ar s fr om 1 4 S S R lo ci
6 T ab le 3 . A lle le s iz es ( bp ) fo r 59 c ul tiv ar s fr om 1 4 S S R lo ci ( C on tin ue d)
TARIM BİLİMLERİ DERGİSİ 2009, Cilt İ 2009, Cilt 15, Sayı 1
A bb re vi at io ns :C S , C ab er ne t S au vi gn on e; M , M er lo t.
Conclusion
Although some plant species from the Mediterranean region of Turkey have been genetically characterized (Bayazit et al. 2007, Ayanoğlu et al. 2007), few studies have explored the genetic diversity within grapevine germplasm of this region. Our findings reported here will assist with future agricultural practices, such as grapevine propagation and breeding and promote better management of regional grapevine cultivars. Importantly, the data reported here can be directly compared with other studies that used these SSR markers on grapes or be integrated into future studies investigating the genetic diversity of grapes from the whole Mediterranean region.
References
Ağaoğlu, Y.S., B. Marasalı ve A. Ergül. 2001. Asmalarda (Vitis vinifera L.) farklı dokulardan izole edilen DNA 'ların RAPD (Random Amplified Polymorphic DNA) tekniğinde kullanımı üzerinde bir araştırma. Tarım Bilimleri Dergisi 7(4): 52-56.
Arroyo-Garcia, R., L. Ruiz-Garcia, L. Boulling, R. Ocete, M. A. López, C. Arnold, A. Ergul, G. Söylemezoğlu, H. I. Uzun, F. Cabello, J. Ibáñez, M. K. Aradhya, A. Atanassov, I. Atanassov, S. Balint, J. L. Cenis, L. Costantini, S. Gorislavets, M. S. Grando, B. Y. Klein, P. McGovern, D. Merdinoglu, I. Pejic, F. Pelsy, N. Primikirios, V. Risovannaya, K. A. Roubelakis-Angelakis, H. Snouss, P. Sotiri, S. Tamhankar, P. This, L. Troshin, J. M. Malpica, F. Lefort and J. M. Martinez-Zapater. 2006. Genetic evidence for the existence of independent domestication events in grapevine. Mol. Ecol. 15: 3707-3714.
Ayanoğlu, H., S. Bayazit, G. İnan, M. Bakır, A.E. Akpınar, K. Kazan and A. Ergül. 2007. AFLP and morphological analyses of genetic diversity in Turkish green plum accessions (Prunus cerasifera L.) adapted to the Mediterranean Region. Sci. Hortic. 114: 263-267. Bayazit, S., K. Kazan, S. Gülbitti, V. Cevik, H. Ayanoğlu and
A. Ergül. 2007. AFLP analysis of genetic diversity in low chill requiring walnut (Juglans regia L.) genotypes from Hatay, Turkey. Scienta Hortic. 111: 394–398.
Bowcook, A. M., A. Ruiz- Linares, J. Tomfohrde, E. Minch, J. R. Kidd and L. L. Cavalli –Sforza. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368: 455-457.
Costantini, L., A. Monaco, J. F. Vouillamoz, M. Forlani and M.S. Grando. 2005. Genetic relationships among local Vitis vinifera cultivars from Campania (Italy). Vitis 44: 25-34.
Dangl, G. S., M. L. Mendum, B. P. Prins, M. A. Walker, C. P. Meredith and C. J. Simon. 2001. Simple sequence repeat analysis of a clonally propagated species: a tool for managing a grape germplasm collection. Genome 44: 432-438.
Ergül, A., K. Kazan, S. Aras, V. Çevik, H. Çelik and G. Söylemezoğlu. 2006. AFLP analysis of genetic variation within the two economically important Anatolian grapevine (Vitis vinifera L.) varietal groups. Genome 49: 467-495.
Fatahi, R., A. Ebadi, N. Bassil, S. A. Mehlenbacher and Z. Zamani. 2003. Characterisation of Iranian grapevine cultivars using microsatellite markers. Vitis 42: 185-192. Lefort, F., M. Lally, D. Thompson and G. C. Douglas. 1998. Morphological traits microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q.robur L.) at Tuallynally, Ireland. Silvae Genet. 47: 5-6.
Martín, J. P., J. Borrego, F. Cabello and J. M. Ortiz. 2003. Characterisation of the Spanish diversity grapevine cultivars using sequence-tagged microsatellite site markers. Genome 46: 1-9.
Minch, E., A. Ruiz-Linares, D. B. Goldstein, M. Feldman and L. L. Cavalli-Sforza. 1995. Microsat (version 1.4d): a computer program for calculating various statistics on microsatellite allele data. Stanford, California, Stanford University.
Nuñez, Y., J. Fresno, V. Torres, F. Ponz and F. J. Gallego. 2004. Practical use of microsatellite markers to manage Vitis vinifera germplasm: Molecular identification of grapevine samples collected blindly in D.O. “El Bierzo” (Spain). J. Hort. Sci. & Biotech. 79: 437-440.
Oraman, M.N. 1969. Arkeolojik buluntuların ışığı altında Türkiye bağcılığının tarihçesi üzerinde araştırmalar (in Turkish). II.Ankara Üniv.Ziraat Fak.Yıllığı 19: 53-75. Paetkau, D., W. Calvert, I. Stirling and C. Strobeck. 1995.
Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4: 347-354.
Rohlf, F. J. 1988. Applied Biostatistics Inc., New York. Santana, J.C., E. Hidalgo, A. I. de Lucas, P. Recio, J. M.
Ortiz, J. P. Martin, J. Yuste, C. Arranz and J. A. Rubio. 2007. Identification and relationships of accessions grown in the grapevine (Vitis vinifera L.) Germplasm Bank of Castilla y Léon (Spain) and the varieties authorized in the VQPRD areas of the region by SSR-marker analysis. Genet. Resour. Crop. Evol. 55: 573– 583.
Sefc, K. M., M. S. Lopes, F. Lefort, R. Botta, K. A. Roubelakis-Angelakis, J. Ibañez, I. Pejic, H.W. Wegner, J. Glössl and H. Steinkellner. 2000. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor. Appl. Genet. 100: 498-505.
Şelli, F., M. Bakır, G. İnan, H. Aygün, Y. Boz, A.S. Yaşasın, C. Özer, B. Akman, G. Söylemezoğlu, K. Kazan and A. Ergül. 2007. Simple sequence repeat-based assessment of genetic diversity in Dimrit and Gemre grapevine accessions from Turkey. Vitis 46:182-187.
8 TARIM BİLİMLERİ DERGİSİ 2009, Cilt 15, Sayı 1
Sneath, P .H. A. and R. R. Sokal. 1973. Numerical taxanomy. San Francisco, CA: Freeman.
Snoussi, H., M. Harbi Ben Slimane, L. Ruiz-Garcia, J. M. Martinez-Zapater and R. Arroyo-Garcia, 2004. Genetic relationship among cultivated and wild grapevine accessions from Tunisia. Genome 47: 1211-1219. This, P., A. Jung, P. Boccacci, J. Borrego, R. Botta, L.
Costantini, M. Crespan, G. S. Dangl, C. Eisenheld, F. Ferreira-Monteiro, S. Grando, J. Ibanez, T. Lacombe, V. Laucou, R. Magalhaes, C. P. Meredith, N. Milani, E. Peterlunger, F. Regner, L. Zulini and E. Maul. 2004. Development of a standard set of microsatellite reference alleles for identification of grape cultivars Theor. Appl. Genet. 109: 1448-1458.
Vouillamoz, J. F., P. E. McGovern, A. Ergül, G. Söylemezoğlu, G. Tevzadze, C. P. Meredith and M. S. Grando. 2006.
Genetic characterization and relationships of traditional grape cultivars from Transcaucasia and Anatolia. Plant Genet. Resour. 4: 144-158.
Wagner, H. W. and K. M., Sefc. 1999. Identity 1.0. Centre for Applied Genetics, University of Agricultural Science, Vienna.
Correspendence Address: Ali ERGÜL
Ankara University, Biotechnology Institute, 06100, Ankara-Turkey
Tel: 0090 312 2225816 Fax : 0090 312 2225872 E-mail: ergul@agri.ankara.edu.tr