Molecular characterization of
Ditylenchus dipsaci on Onion
in Turkey
E. Yavuzaslanoglu
&O. Ates Sonmezoglu
&N. Genc
&Z. Akar
&B. Terzi
Accepted: 11 October 2017 / Published online: 16 October 2017 # Koninklijke Nederlandse Planteziektenkundige Vereniging 2017
Abstract Ditylenchus dipsaci is a species complex
in-cluding diploid and polyploid individuals. The onion
race of D. dipsaci is a sensu stricto group and has a
wide range of host spectrum. Identification of the
D. dipsaci onion race is difficult using morphological
and morphometrical methods. Species specific primers
are mostly used in molecular approaches for
identifica-tion of D. dipsaci populaidentifica-tions. Fifty one
morphological-ly selected Ditylenchus spp. populations from onion
production areas in Turkey were subjected to molecular
identification using four D. dipsaci species specific
primer sets (PF1-PR1, PF2-PR2, DdpS1-rDNA2,
DitNF1- rDNA2, H05-H06) targeting 5.8S and 18S
rDNA, ITS1 and flanking ITS regions. Thirty nine
percent of the nematode samples were positive with four
primers tested, while four of the nematode samples gave
specific bands with H05-H06 primers. Ditylenchus
dipsaci sensu stricto was identified with specific primer
sets in Adana, Hatay, Tekirdag, Bursa, Aksaray,
Karaman, Eskisehir and Ankara provinces in
Mediter-ranean, Trace, Aegean and Central Regions in Turkey.
Keywords Onion . Stem and bulb nematode .
Species specific primer . Molecular characterization
Ditylenchus dipsaci (Kühn 1857) is among the
signifi-cant damaging plant parasitic nematodes on agricultural
production in temperate conditions. Ditylenchus dipsaci
has very high intraspecific variation (Subbotin et al.
2005
). It has more than 30 races multiplying on 500
plant species (Sturhan and Brzenski
1991
). Therefore, it
is named as species complex. Last taxonomical studies
divided D. dipsaci species complex into two groups;
one is
BD. dipsaci sensu stricto^ group including
dip-loid members, the other group includes polypdip-loid
mem-bers, which is also subdivided into 6 groups;
Ditylenchus sp. B from Vicia faba, Ditylenchus sp. C
from Cirsium setosum, Ditylenchus sp. D related to
Pilosella genera plants, Ditylenchus sp. E found on
Crepis praemorsa, Ditylenchus sp. F related to
Pilosella and Leontodon genera plants and Ditylenchus
sp. G subgroup identified on Plantago maritima plant
(Subbotin et al.
2005
). Further studies identified
Ditylenchus sp. C as Ditylenchus weischeri (Chizhov
et al.
2010
) and Ditylenchus sp. B group from Vicia
faba as Ditylenchus gigas (Vovlas et al.
2011
). Diploid
nematode populations showed very close phylogenetic
relationships and were classified as races (Subbotin
et al.
2005
). Janssen (
1994
) described eight races from
https://doi.org/10.1007/s10658-017-1366-7
E. Yavuzaslanoglu (*)
Department of Plant and Animal Production, Technical Sciences Vocational School, Karamanoğlu Mehmetbey University, Karaman, Turkey
e-mail: eyavuzaslanoglu@kmu.edu.tr O. Ates Sonmezoglu (*)
:
B. TerziDepartment of Bioengineering, Faculty of Engineering, Karamanoglu Mehmetbey University, Karaman, Turkey e-mail: ozlemsonmezoglu@kmu.edu.tr
N. Genc
:
Z. AkarInstitute of Science, Karamanoglu Mehmetbey University, Karaman, Turkey
Medicago sativa, Trifolium pratense, Avena sativa,
Secale cereale, Beta vulgaris, Narcissus spp., Tulipa
spp. and Allium cepa. The onion race of D. dipsaci
was commonly identified by researchers to damage
onion, garlic, pea and bean, but other hosts have
differ-entiating reactions in different studies (Janssen
1994
;
Mennan
2001
). Due to the inconsistency over host
status, control of the nematode using host crop rotation
is difficult. Anabiosis ability of the nematode in the
absence of the host plant makes it also difficult to
control the nematode under agricultural practices.
Host preferences of the nematodes could be
deter-mined by genetically using techniques determining
polymorphism on pathogenicity genes. The
complex-ity and difficulty on determination of host status and
morphological similarities lead to development of
dif-ferent molecular taxonomic approaches for difdif-ferenti-
differenti-ation of the D. dipsaci species complex on different
host plants (Esquibet et al.
1998
; Subbotin et al.
2005
;
Kerkoud et al.
2007
; Zouhar et al.
2007
; Douda et al.
2013
; Jeszke et al.
2014
). Polymorphism on the
inter-nal transcribed spacer (ITS) regions of ribosomal
DNA is the most commonly used approach for
deter-mination of intraspecific variations of D. dipsaci
(Esquibet et al.
2003
; Marek et al.
2005
; Subbotin
et al.
2005
; Kerkoud et al.
2007
; Zouhar et al.
2007
).
Ditylenchus dipsaci has a local distribution in
Eu-rope being listed on the quarantine A2 list by the
European and Mediterranean Plant Protection
Organi-zation (EPPO) (EPPO
2017
). It was first identified on
onion by Yuksel (
1958
) in Turkey. It was recorded on
onion-growing areas in Trace, Central Anatolian
Pla-teau and Black Sea Region in Turkey (Saltukoglu
1974
; Ozturk
1990
; Mennan and Ecevit
2002
;
Yavuzaslanoglu et al.
2015a
). Yield losses of 41.5–
65% on onion was determined in Turkey (Mennan and
Ecevit
2002
; Yavuzaslanoglu et al.
2015b
).
Studies of the distribution of D. dipsaci on onion
in Turkey to-date have used morphological and
mor-phometric techniques. There is no available detailed
information on the species complex distribution in
large scale onion growing areas of Turkey. The aim
of this study was to investigate the D. dipsaci sensu
stricto group distribution on onion, using specific
PCR techniques, for the market-scale onion
produc-tion areas in Turkey.
Materials and methods
Nematode populations
Nematode populations were obtained from onion
fields in ten provinces in Mediterranean, Trace,
Ae-gean and Central Anatolia Regions in Turkey on
April to May 2016. In total, 51 nematode populations
were studied; five from Adana, six from Hatay, seven
from Tekirdag, one from Balikesir, nine from Bursa,
one from Aksaray, six from Karaman, two from
Kon-ya, twelve from Ankara, and two from Eskisehir
provinces where onion production is most practised
in Turkey (Table
2
). The nematodes used for
molec-ular identification were extracted from either plant
tissues or soil using a
‘modified baerman funnel’
technique (Hooper
1986
). Nematodes were identified
morphologically at genus level as Ditylenchus spp.
and collected with a pasteur pipette from samples in a
PCR tube.
Molecular characterization
DNA was extracted from each nematode population
according to Holterman et al. (
2006
) with some
modifications. Five to ten individual nematodes
were transferred with 25
μl sterile distilled water
into an Eppendorf tube and homogenized in 25
μl
lysis buffer (WLB
+). WLB+ contained 10
μl
beta-merkaptoetanol, 40
μl 20 mg/ml Proteinase K and
950
μl WLB- buffer (2 ml 1 M NaCl, 2 ml 1 M
Tris-HCl and 5.5 ml of ddH
2O). The mixture was
incu-bated for 90 min at 65 °C and finally denatured
5 min at 95 °C. The tubes were centrifuged at
14,000 rpm for 1 min and stored at
−20 °C. DNA
was eluted in 20
μL ddH
2O and stored at
−20 °C.
PCR reactions were carried out as follows: an initial
denaturation step of 3 min at 94 °C was followed by
37 cycles of denaturation for 1 min at 94 °C, annealing
for 45 s at 55
–62 °C (depending upon the annealing
temperature of the primers), extension for 2 min at
72 °C, and conclusion with a final extension step for
10 min at 72 °C. Negative control samples containing
only sterile distilled water (no DNA target) were included.
DNA from a morphologically identified D. dipsaci culture
(Yavuzaslanoglu et al.
2015a
) was used as a positive
control. Specific amplifications were repeated three times
to assess the reproducibility of PCR. PCR amplicons were
separated in 1–2% agarose gels. Electrophoresis was
con-ducted at 90 V of constant power for 3 h.
PCR reactions (for all primers except PF1-PR1
prim-er set) wprim-ere carried out in a BIO-RAD (C1000 Touch)
thermal cycler in a total volume of 25
μL using 1.5 U
Taq DNA Polymerase (Thermo), 200
μM each dNTPs,
1.2
μM MgCl
2, 0.5
μM each primer, 10 x Tag Buffer
with KCL. PCR amplification for PF1-PR1 primer was
determined in a total reaction volume of 25
μL, with
2 ng DNA as template, 7.5
μL Dream Taq Green MM
(Thermo), 0.5
μM each primer.
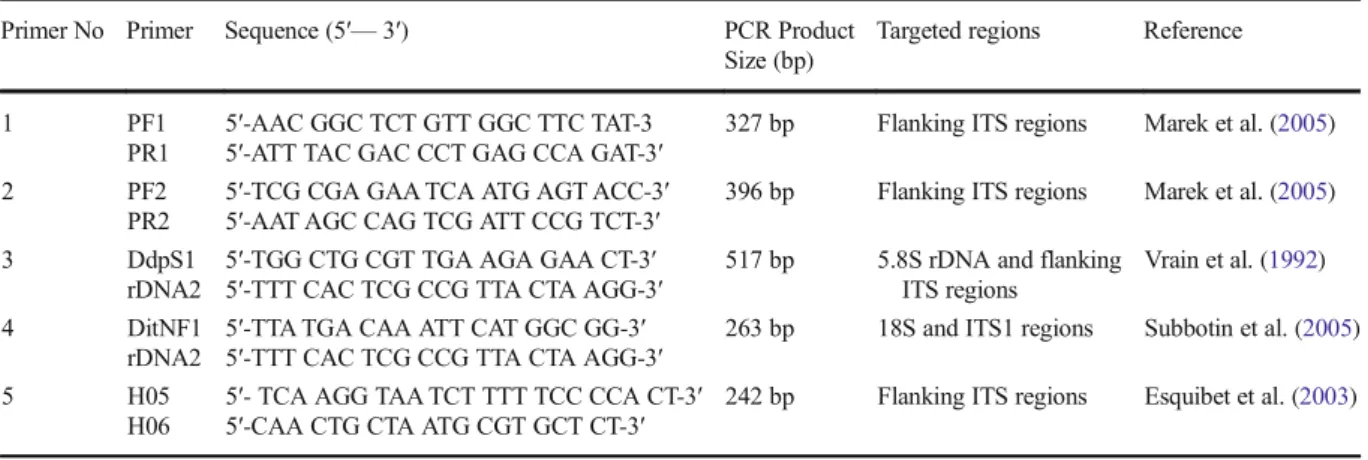
The five species-specific PCR primers targeted to
amplify 5.8S, 18S rDNA genes, ITS1 and flanking
ITS regions used for the identification of D. dipsaci
were listed on Table
1
.
Results and discussion
Using the species-specific PCR primers and nematode
DNA extracted from onion plants and soils, PCR
frag-ments were successfully amplified at expected product
size for each primer set in D. dipsaci samples. All of the
primers give present or absent a specific size band in
D. dipsaci. The used primers in the study were
ampli-fying only D. dipsaci sensu stricto group.
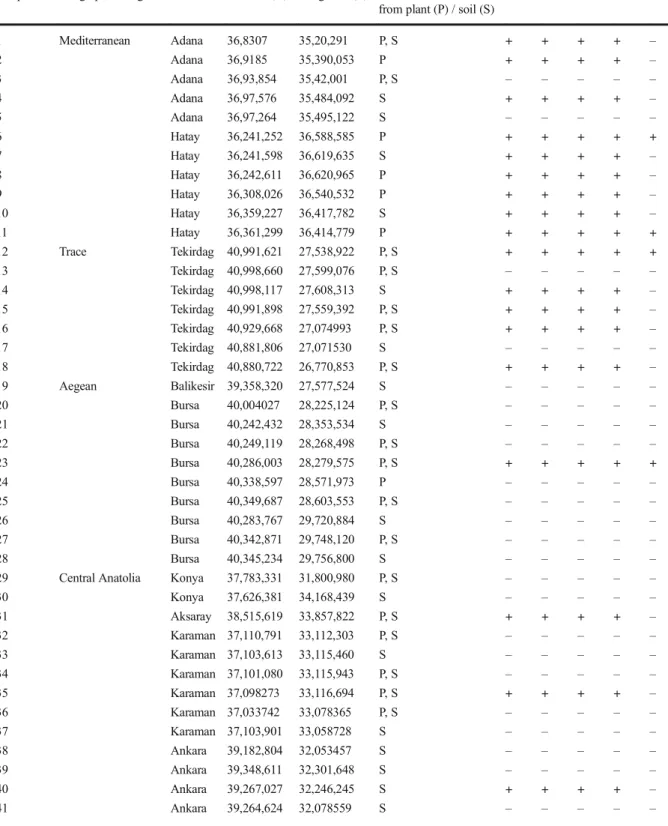
Fifteen of the 51 nematode populations showed
ex-pected band size with all primer sets used in the
molec-ular screening. The numbers of these samples are 1, 2
and 4 in Adana Province; 8 and 11 in Hatay Province;
12, 15 and 18 in Tekirdag Province; 25 in Bursa
Prov-ince; 31 in Aksaray ProvProv-ince; 34 and 35 in Karaman
Province; 50 in Eskisehir Province; 54 in Amasya
Prov-ince and 58 in Corum ProvProv-ince. Twenty one of the
samples were negative with all primer sets. Twenty of
the samples gave positive reaction with at least one of
the primers, reactions were differentiating among
primers (Table
2
).
The amplicon of PF1-PR1 primer set targeting
flanking ITS spacer regions was 327 bp for studied
D. dipsaci populations and 20 of the nematode
popula-tions produced specific bands of D. dipsaci sensu
stricto. Similarly, a fragment with 396 bp band size
was amplified with 20 nematode populations using
PF2-PR2 primers. For both primer sets, samples not
including D. dipsaci observed no amplification in
ac-cordance with Marek et al. (
2005
).
Expected PCR product size 517 bp (Vrain et al.
1992
) for D. dipsaci were obtained using
DdpS1-rDNA2 primers targeted 5.8S rDNA gene and flanking
ITS regions. The specific amplicon was observed for
twenty samples.
The amplicon of DitNF1-rDNA2 primer set is 263 bp
for D. dipsaci sensu stricto (Subbotin et al.
2005
). The
primers do not amplify for D. dipsaci giant race, D.
myceliophagus and Ditylenchus sp. Twenty of the tested
populations were found to be D. dipsaci sensu stricto
using this primer set.
The used primer sets can be used successfully for
D. dipsaci specific identification. Total 20 samples
Table 1 SCAR and SSR primers used in identification of D. dipsaci nematodes Primer No Primer Sequence (5′— 3′) PCR Product
Size (bp)
Targeted regions Reference
1 PF1 5′-AAC GGC TCT GTT GGC TTC TAT-3 327 bp Flanking ITS regions Marek et al. (2005) PR1 5′-ATT TAC GAC CCT GAG CCA GAT-3′
2 PF2 5′-TCG CGA GAA TCA ATG AGT ACC-3′ 396 bp Flanking ITS regions Marek et al. (2005) PR2 5′-AAT AGC CAG TCG ATT CCG TCT-3′
3 DdpS1 5′-TGG CTG CGT TGA AGA GAA CT-3′ 517 bp 5.8S rDNA and flanking ITS regions
Vrain et al. (1992) rDNA2 5′-TTT CAC TCG CCG TTA CTA AGG-3′
4 DitNF1 5′-TTA TGA CAA ATT CAT GGC GG-3′ 263 bp 18S and ITS1 regions Subbotin et al. (2005) rDNA2 5′-TTT CAC TCG CCG TTA CTA AGG-3′
5 H05 5′- TCA AGG TAA TCT TTT TCC CCA CT-3′ 242 bp Flanking ITS regions Esquibet et al. (2003) H06 5′-CAA CTG CTA ATG CGT GCT CT-3′
Table 2 Origin and extraction medium of nematode populations tested in the study and PCR results with specific primer sets (set 1: PF1-PR1, set 2: PF2-PR2, set 3: DdPS1-rDNA2, set 4: DitFN1-rDNA2, set 5: H05-H06)
Sample no Geographical region Province Latitude (N) Longitude (E) Nematode extracted from plant (P) / soil (S)
Set 1 Set 2 Set 3 Set 4 Set 5
1 Mediterranean Adana 36,8307 35,20,291 P, S + + + + – 2 Adana 36,9185 35,390,053 P + + + + – 3 Adana 36,93,854 35,42,001 P, S – – – – – 4 Adana 36,97,576 35,484,092 S + + + + – 5 Adana 36,97,264 35,495,122 S – – – – – 6 Hatay 36,241,252 36,588,585 P + + + + + 7 Hatay 36,241,598 36,619,635 S + + + + – 8 Hatay 36,242,611 36,620,965 P + + + + – 9 Hatay 36,308,026 36,540,532 P + + + + – 10 Hatay 36,359,227 36,417,782 S + + + + – 11 Hatay 36,361,299 36,414,779 P + + + + + 12 Trace Tekirdag 40,991,621 27,538,922 P, S + + + + + 13 Tekirdag 40,998,660 27,599,076 P, S – – – – – 14 Tekirdag 40,998,117 27,608,313 S + + + + – 15 Tekirdag 40,991,898 27,559,392 P, S + + + + – 16 Tekirdag 40,929,668 27,074993 P, S + + + + – 17 Tekirdag 40,881,806 27,071530 S – – – – – 18 Tekirdag 40,880,722 26,770,853 P, S + + + + – 19 Aegean Balikesir 39,358,320 27,577,524 S – – – – – 20 Bursa 40,004027 28,225,124 P, S – – – – – 21 Bursa 40,242,432 28,353,534 S – – – – – 22 Bursa 40,249,119 28,268,498 P, S – – – – – 23 Bursa 40,286,003 28,279,575 P, S + + + + + 24 Bursa 40,338,597 28,571,973 P – – – – – 25 Bursa 40,349,687 28,603,553 P, S – – – – – 26 Bursa 40,283,767 29,720,884 S – – – – – 27 Bursa 40,342,871 29,748,120 P, S – – – – – 28 Bursa 40,345,234 29,756,800 S – – – – – 29 Central Anatolia Konya 37,783,331 31,800,980 P, S – – – – – 30 Konya 37,626,381 34,168,439 S – – – – – 31 Aksaray 38,515,619 33,857,822 P, S + + + + – 32 Karaman 37,110,791 33,112,303 P, S – – – – – 33 Karaman 37,103,613 33,115,460 S – – – – – 34 Karaman 37,101,080 33,115,943 P, S – – – – – 35 Karaman 37,098273 33,116,694 P, S + + + + – 36 Karaman 37,033742 33,078365 P, S – – – – – 37 Karaman 37,103,901 33,058728 S – – – – – 38 Ankara 39,182,804 32,053457 S – – – – – 39 Ankara 39,348,611 32,301,648 S – – – – – 40 Ankara 39,267,027 32,246,245 S + + + + – 41 Ankara 39,264,624 32,078559 S – – – – –
(39%) among the 51 nematode samples from onion
plants and soils were commonly identified as
D. dipsaci sensu stricto by molecular characterization.
Nematodes from 31 locations (61%) did not produced
specific bands with any of the primer sets tested.
How-ever, a specific band for at least one of the primer sets
used was observed for 20 nematode samples (39%).
Duo to the complex nature of the D. dipsaci species
complex, it is very useful to use molecular techniques
for the routine identification of the D. dipsaci samples
from different host plants. Vrain et al. (
1992
), Marek
et al. (
2005
) and Subbotin et al. (
2005
) used and
recommended the specific primers to identify
D. dipsaci sensu stricto. It has also been reported that
rDNA ITS regions can be successfully used for
phylo-genetic analyzes (Subbotin et al.
2005
; Marek et al.
2010
; Vovlas et al.
2011
; Pethybridge et al.
2016
). The
precise identification of the nematode is an important
step for effective control of the host plant. For this
purpose, to know the distribution of the sensu stricto
group of D. dipsaci including the onion race on onion
plants and production soils in Turkey is very valuable
and it is first report on distribution of D. dipsaci sensu
stricto in onion-growing areas in Turkey using
molec-ular tools.
Acknowledgements This research has been financially support-ed by Turkish Scientific and Technical Research Council (TUBITAK) (Project No: 215O468).
Compliance with ethical standards The manuscript has not been submitted to any other journal. The manuscript has not been
published previously (partly or in full). Preliminary data was pre-sented in 32nd ESN Symposium, held in Braga, Portugal, in 28th August -1st September 2016;BMolecular identification of stem and bulb nematode (Ditylenchus dipsaci) on onion in Turkey^ by Nimet Genc, Ozlem Sonmezoglu and Elif Yavuzaslanoglu. A single study was not split up into several parts to increase the quantity of submissions and submitted to various journals or to one journal over time. No data have been fabricated or manipulated (including images) to support our conclusions. No data, text or theories by others are presented.
The research does not require any ethical council permission. Consent to submit has been received explicitly from all co-authors, before the work is submitted. Authors whose names appear on the submission have contributed sufficiently to the scientific work and therefore share collective responsibility and accountability for the results.
References
Chizhov, V. N., Borisov, B. A., & Subbotin, S. A. (2010). A new stem nematode, Ditylenchus weischeri sp. n. (Nematoda: Tylenchida), a parasite of Cirsium arvense (L.) Scop. in the Central Region of the Non-Chernozem Zone of Russia. Russian Journal of Nematology, 18(2), 95.
Douda, O., Marek, M., Zouhar, M., & Rysanek, P. (2013). Insights into the structure and phylogeny of the 28S rRNA expansion segments D2 and D3 of the plant-infecting nematodes from the genus Ditylenchus (Nematoda: Anguinidae). Phytopathologia Mediterranea, 52(1), 84–97.
EPPO (2017). A2 List of pests recommended for regulation as quarantine pests (version 2013–09). http://www.eppo. int/QUARANTINE/listA2.htm. Accessed 1 Feb 2017. Esquibet, M., Bekal, S., Castagnone-Sereno, P., Gauthier, J.-P.,
Rivoal, R., & Caubel, G. (1998). Differentiation of normal and giant Vicia faba populations of the stem nematode Table 2 (continued)
Sample no Geographical region Province Latitude (N) Longitude (E) Nematode extracted from plant (P) / soil (S)
Set 1 Set 2 Set 3 Set 4 Set 5
42 Ankara 39,277,253 32,075229 S – – – – – 43 Ankara 39,346,711 31,979,551 S – – – – – 44 Ankara 39,434,225 31,985,016 S – – – – – 45 Ankara 39,719,689 32,393,649 S – – – – – 46 Ankara 39,707,854 32,403,380 S – – – – – 47 Ankara 39,838,486 32,293,308 S – – – – – 48 Ankara 39,846,889 32,298,189 S – – – – – 49 Ankara 40,110,527 31,857,132 S – – – – – 50 Eskisehir 39,707,117 30,412,748 S + + + + – 51 Eskisehir 39,458,774 31,328,988 S + + + + –
Ditylenchus dipsaci: agreement between RAPD and pheno-typic characteristics. Heredity, 81(3), 291–298.
Esquibet, M., Grenier, E., Plantard, O., Andaloussi, A., & Caubel, G. (2003). DNA polymorphism in the stem nematode Ditylenchus dipsaci: Development of diagnostic markers for normal and giant races. Genome, 46, 1077–1083. Holterman, M., Wurff, A., Elsen, S., Megen, H., Bongers, T.,
Holovachov, O., et al. (2006). Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution towards crown clades. Molecular Biology and Evolution, 23(9), 1792–1800. Hooper, D. J. (1986). Extraction of Free Living Stages from Soil. In
J. F. Southey (Ed.), Laboratory methods for work with plant and soil nematodes. London: Her Majesty’s Stationery Office. Janssen, G. J. W. (1994). The relevance of races in Ditylenchus dipsaci (Kühn) Filipjev, the stem nematode. Fundamental and Applied Nematology, 17(5), 469–473.
Jeszke, A., Budziszewska, M., Dobosz, R., Stachowiak, A., Protasewicz, D., Wieczorek, P., et al. (2014). A compar-ative and phylogenetic study of the Ditylenchus dipsaci, Ditylenchus destructor and Ditylenchus gigas popula-tions occurring in Poland. Journal of Phytopathology, 162(1), 61–67.
Kerkoud, M., Esquibet, M., Plantard, O., Avrillon, M., Guimier, C., Franck, M., et al. (2007). Identification of Ditylenchus species associated with fabaceae seeds based on a specific polymerase chain reaction of ribosomal DNA-ITS regions. European Journal of Plant Pathology, 118(4), 323–332. Marek, M., Zouhar, M., Rysanek, P., & Havranek, P. (2005).
Analysis of ITS sequences of nuclear rDNA and develop-ment of a PCR-based assay for the rapid identification of the stem nematode Ditylenchus dipsaci (Nematoda: Anguinidae) in plant tissues. Helminthologia, 42(2), 49.
Marek, M., Zouhar, M., Douda, O., & Mazakova, J. v. R. (2010). Bioinformatics-assisted characterization of the ITS1-5-8S-ITS2 segments of nuclear rRNA gene clusters and its exploi-tation in molecular diagnostics of European crop-parasitic nematodes of the genus Ditylenchus. Plant Pathology, 59(5), 931–943.
Mennan, S. (2001). Amasya Suluova İlçesi Soğan Ekim Alanlarında Soğan Sak Nematodu Ditylenchus dipsaci (Kühn, 1857) (Nematoda: Tylenchida: Anguinidae) Populasyonunun Bitki Koruma Yönünden Araştırılması. Samsun: Ondokuz Mayıs University PhD Thesis.
Mennan, S., & Ecevit, O. (2002). Farklı Preparatların Ditylenchus dipsaci Soğan Irkına Karşı Etkinliği Üzerinde Araştırmalar. Ondokuz Mayıs Üniversitesi Ziraat Fakültesi Dergisi, 17(16), 20–24.
Ozturk, G. (1990). Taxonomic studies on plant parasitic nema-todes in Tylenchida order in Konya, Karaman ve Nevsehir provinces onion (Allium cepa L.) planting areas. Ankara: Ankara University PhD Thesis.
Pethybridge, S. J., Gorny, A., Hoogland, T., Jones, L., Hay, F., Smart, C., et al. (2016). Identification and characterization of Ditylenchus Spp. populations from garlic in New York State, USA. Tropical Plant Pathology, 41(3), 193–197.
Saltukoglu, M.E. (1974). A taxonomical and morphological study of tylenchida (Nematoda) from the Istanbul Area (Turkey). Belgium: State University of Gent PhD Thesis.
Sturhan, D., & Brzenski, M. W. (1991). Stem and bulb nem-atodes, Ditylenchus spp. In W. R. Nickle (Ed.), Manual of agricultural nematology (pp. 423–465). New York: Marcel Dekker Publications.
Subbotin, S. A., Madani, M., Krall, E., Sturhan, D., & Moens, M. (2005). Molecular diagnostics, taxonomy and phylogeny of the stem nematode Ditylenchus dipsaci species complex based on the sequences of the intertranscribed spacer-rDNA. Phytopathology, 95, 1308–1315.
Vovlas, N., Troccoli, A., Palomares-Rius, J. E., De Luca, F., Liebanas, G., Landa, B. B., et al. (2011). Ditylenchus gigas n. sp. parasiting broad bean: a new stem nematode singled out from the Ditylenchus dipsaci species complex using a polyphasic approach with molecular phylogeny. Plant Pathology, 60, 762–775.
Vrain, T. S., Wakarchuk, D. A., Levesque, A. C., & Hamilton, R. I. (1992). Interspecific rDNA restriction fragment length poly-morphism in The Xiphinema Americanum group. Fundamental and Applied Nematology, 15(16), 563–573. Yavuzaslanoglu, E., Dikici, A., & Elekcioglu, I. H. (2015a). Effect
of Ditylenchus dipsaci Kühn, 1857 (Tylenchida: Anguinidae) on onion yield in Karaman Province, Turkey. Turkish Journal of Agriculture and Forestry, 39(2), 227–233. Yavuzaslanoglu, E., Dikici, A., Elekcioglu, I. H., & Aydogdu, M.
(2015b). Distribution of nematodes on onion and their rela-tionship with soil physicochemical characteristics in Karaman province, Turkey. Turkish Journal of Entomology, 39(3), 251–259.
Yuksel, H. S. (1958). First detection of stem and bulb nematode (Ditylenchus dipsaci grup) on onions on Central Anatolian Plateau in Turkey. Tomurcuk, 77, 5–6.
Zouhar, M., Marek, M., Douda, O., Mazakova, J., & Rysanek, P. (2007). Conversion of sequence-characterized amplified re-gion (SCAR) bands into high-throughput DNA markers based on RAPD technique for detection of the stem nematode Ditylenchus dipsaci in crucial plant hosts. Plant Soil Environment, 53(3), 97.