INVESTIGATION OF SOME PROPERTIES OF BACILLUS SPP.
ISOLATED FROM ARCHAEOLOGICAL EXCAVATIONS SOIL
Yasemin Yeşiltaş1, Ferdağ Çolak1,* and Elif Genç2
1 Department of Biology, Faculty of Arts & Science, University of Dumlupinar, Kütahya, Turkey 2Department of Archaeology, Faculty of Science and Letters, Çukurova University, Adana, Turkey
ABSTRACT
In this study, soil samples were taken from the Kuriki Mound, the silo, the furnace ruins, and graves within the excavation area, 14 km south of the city of Batman. VITEK biochemical identification tests determined that there were 8 isolates of Brevibacillus borstelensis, 7 isolates of
Bacil-lus cereus, 6 isolates of B. subtilis, 1 isolate of Paenibacil-lus macerans, 1 isolate of B. popillia, 1 isolate of B. poly-myxa isolates, 2 isolates of B. coagulans, and 1 isolate of B. larvae. The identified strains were subjected to heavy
metals and antibiotic resistance profiling, antimicrobial and enzyme activity, and PHB formations were investi-gated. The highest heavy metal resistance in terms of Min-imal Inhibitory Concentration (MIC) value was observed against the manganese (16Mm), in comparison to the ref-erence strain (B. subtilis NRRL B-209). The highest level of antibiotic resistance was observed against cefotaxime, while the lowest antibiotic resistance was exhibited against gentamycin. Bacillus spp., antimicrobial activity against Gram-negative/positive bacteria, yeast, mold and clinical isolates was determined using the agar well diffusion tech-nique. The tested isolates were devoid of any antimicrobial activity against Gram negative bacteria. The bacilli ob-tained in the course of our study showed activity against the isolates B. cereus, S. epidermidis and B. subtilis. Clini-cal isolates showed maximum antimicrobial activity against Acinetobacter spp., while against Vancomycine re-sistant Enterecocci (VRE) they showed no antimicrobial activity. The bacillus Bacillus isolates showed antifungal activity against the previously mentioned yeasts
(Saccho-romyces cerevisia, S. baulardi and Rhodotorula. rubra).
10% of the Bacillus isolates showed amylase and protease activity. Poly-ß hydroxybutyrate (PHB) was observed in only 4 out of the 27 Bacillus isolates.
KEYWORDS: Antimicrobial activity, archaeology, Bacillus spp,
en-zymes, heavy metal, PHB.
* Corresponding author
1. INTRODUCTION
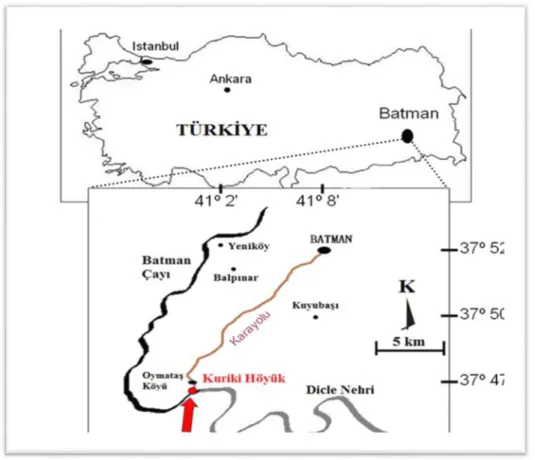
The Kuriki Mound is located near the village of Oymataş, 14 kilometers south of the city of Batman, on the left bank of the Batman River. The mound is located at the confluence of the River Tigris and the Batman River, within the rivers’ terrace lands, which are suitable for agri-cultural production. Two main areas of occupation have been distinguished and named Kuriki Mound 1 and Kuriki Mound 2, as shown in Figure 1. The first season of archae-ological excavation at Kuriki Mound was conducted in 2009, as part of the ILISU and hydroelectric power plant (HES) Projects. The settlement contained four levels, which were subdivided into twelve phases, dating back about 5000 years. The first occupation documented at the site dates back to the end of the 4th/beginning of the 3rd millennia. After a long period of abandonment, Kuriki was re-occupied during the 1st millennium BC, and the latest occupation occurred during the late 1st millennium AD [1-4]. .
Spores of various Bacillus species are formed in spor-ulation, a process that is generally induced by reduced lev-els of nutrients in the environment. The formation of a spore generates a cell type that can survive for extended periods with little or no nutrients, yet is poised to return to life if nutrients become available. As spores may have to survive for long periods without nutrients, they are meta-bolically dormant, contain little or no high energy com-pounds such as ATP and NADH, exhibit no detectable me-tabolism of endogenous or exogenous compounds and lit-tle, if any, enzyme activity in the spore core, the analogue of the protoplast of a growing cell [5].
Heavy metals as natural components of the earth’s crust are increasingly found in microbial habitat due to several natural and anthropogenic processes. However, microbes have evolved mechanisms to tolerate the presence of heavy metals either by efflux, complexation or reduction of metal ions or to use them as terminal electron acceptors in anaero-bic respiration [6, 7]. It has been shown that a correlation exists between metal tolerance and antibiotic resistance in bacteria because of the likelihood that resistance genes to both (antibiotics and heavy metals) may be closely located on the same plasmid in bacteria and the presence of the or-ganisms that possess specific mechanisms of resistance to
FIGURE 1 - Kuriki mound location map.
heavy metals increases destruction or transformation of toxic substances in the natural environment. Consequently, the range of genes carried on these plasmids (frequently asso-ciated with these heavy metal resistant determinants) was shown to extend far beyond those coding for antibiotic re-sistance [8].
Heavy metals are stable and persistent environmental contaminants since they cannot be degraded or destroyed. Therefore, they tend to accumulate in soils and sediments. While some heavy metals are required in trace amounts as nutrients, they become strongly inhibitory for microorgan-isms at relatively low concentrations. Toxicity occurs through the displacement of essential metals from their na-tive binding sites or through ligand interactions. Toxicity results from alterations in the conformational structure of nucleic acids and proteins and interference with oxidative phosphorylation and osmotic balance. To survive under metal-stressed conditions, bacteria have evolved several types of adaptation mechanisms to tolerate the uptake of heavy metal ions. These mechanisms include the efflux of metal ions outside the cell, accumulation and complexation of the metal ions inside the cell, and the reduction of the heavy metal ions to a less toxic state. Bacteria have adapted to heavy metals through a variety of chromosomal-, trans-poson-, and plasmid-mediated resistance systems [9].
PHB is well known as a carbon and energy reserve pro-duced by a variety of microorganisms and its synthesis is
favored by environmental stresses, such as nitrogen, phos-phate or oxygen limitation. PHB and other PHAs are syn-thesized and deposited intracellularly in the form of gran-ules and might amount up to 90% of the cellular dry weight. Accumulation of intracellular storage polymers has been considered a strategy used by bacteria to increase sur-vival in a changing environment [10].
Of all the known PHAs, poly- 3-hydroxybutyrate, PHB, is the most commonly and widely produced homo-polymer by many bacteria. Current applications of PHB-based polymers or composites include the packaging in-dustry, medicine, pharmacy, agriculture, food inin-dustry, raw material for enantiomerically pure chemicals and the paint industry [11].
The Bacillus species that produce antibiotics are B.
subtilis, B. polymyxa, B. brevis, B. licheniformis, B. circu-lans and B. cereus. The majority of studied antibiotics
pro-duced by Bacillus strains are polypeptides of low molecular weight that are synthesized by ribosomal or nonribosomal mechanisms [12]. Polypeptide antibiotics produced by
Ba-cillus that are used in medical treatments are bacitracin,
gramicidins, polymyxin, and tyrotricidin [13]. B. cereus and some closely related species from the genus Bacillus have several features including the production of various biologi-cally active metabolites, i.e. antibiotics, proteinases and bacteriocins. It is well-known that most, if not all, bacterial species are capable of producing a heterogeneous array of
molecules in the course of their growth in vitro (and pre-sumably also in their natural habitats) that may be inhibi-tory to other bacteria [14].
Therefore, we have designed this study with the aim of isolating endospore forming bacilli from Kuriki mound. Further, our aim was to characterize the isolated bacteria for their antibiotic and heavy metal resistance pattern as well as enzyme activities and PHB formation.
2. MATERIALS AND METHODS
2.1 Isolation and Identification of Bacillus strains
Soil samples were taken from excavated soil at three different locations (silo, furnace ruins, and graves from the soil), at Kuriki mound. An impressive monumental build-ing of the Level II, datbuild-ing to the end of the 1st millennium BC/beginning of the 1st millennium AD, was excavated on the top of the mound. This building was centered on a long corridor overlooked by two pairs of three rooms. In order to establish a stratigraphic sequence of the settlement, a deep sounding was dug. The main soil was reached by dig-ging as far as five meters down from the corridor, and here the remains of a silo were found and containing a huge amount of carbonized lentils and wheat. A cist tomb made by slab stones containing a 5-6 month [1-4].
Ten grams of soil sample was serially diluted in sterile water and dilutions were surface-plated on Nutrient agar. The soils were pasteurized at 80ºC for 10 minutes to select the Gram positive spore forming bacteria, Bacillus spp. Af-ter spreading 100 μL of each suspension on Nutrient Agar (NA), the sample was aerobically incubated for 2 days at 37°C. After the incubation periods, colonies were selected for isolation. The purified isolate was maintained on agar medium, and also stored as glycerol (20%, v/v) stocks at -18° C.
The morphological and some biochemical properties of the isolate were determined by carrying out Gram stain, endospore stain, catalase test, anaerobic growth, voges prouskauer test, hydrolysis of casein, gelatin, and starch, growth at different pH, temperature, and NaCl concentra-tions in accordance with Bergey’s Manual of Systematic Bacteriology [15]. The bacteria were identified using a VI-TEK Compact 1 System based on the differences of carbon source utilization.
2.2 Determination of Minimum Inhibitory Concentrations (MIC) of heavy metals
The Minimal Inhibitory Concentration (MIC) of the metals for the isolate was determined by the plate dilution method, as adopted by Alam and Malik [16] and Alam et al.[17]. The MIC for two bacterial isolates in terms of seven heavy metals was determined using NA containing Ni2+, Zn2+, Co2+, Fe 3+, Cr6+, Pb2+, Mn2+, Cu2+ and Cd +2 at concentrations ranging from 0.0625mM/mL to 16mM/mL. The metals were added as Ni(CH3COO)2.4H2O, Zn(CH3COO)2.H2O, Co(CH3COO)2.4H2O, FeSO4.7H2O,
CrCl36H2O, PbCl2, MnCl2.4H2O, Cu(CH3COO)2.H2O and CdCl2.4H2O. Stock solutions of the metal salts were pre-pared in sterile distilled water. The solutions were then sterilized by filtration with syringe type filters (with a pore diameter of 0.22 μm) and were added to the NA plates in various concentrations which were then spot-inoculated with approximately 108CFU/mL. The density of this cul-ture was adjusted to 0.5 McFarland (at 625nm, 0.08-0.1 ab-sorbance). The plates were incubated at 37°C for 24 hours. The lowest concentration of metal inhibiting the growth of the microorganisms was considered as the MIC of the metal against the strain tested. The isolates were consid-ered resistant if the MIC values exceeded that of the B.
sub-tilis NRRL B-209 strain, which was used as the control.
2.3 Sensitivity to the Antibiotics
Bacillus isolates were cultured in Nutrient Broth (NB)
medium and incubated at 37°C for 24 hours, and a suspen-sion containing 108CFU/mL for each isolate was prepared. 100μL of each suspension was spread on the plates con-taining Mueller-Hinton Agar (MHA) medium (15 mL). Antibiotic discs, namely, amikacin (AK 30µg), gentamicin (CN 30µg), tetracycline (TE 30µg), cefotaxime (CTX 30µg), vancomycin (VA 30µg), oflaxacin (OFX 5µg), sulbactam (SAM 30µg), chloramphenicol (C 30µg), oxa-cillin (OX 1µg), ampicilin (AMP 10µg), clindamicin (DA 10µg), streptomicin (S 10µg), meticilin (Met 10µg), kana-mycine (K 30µg), imipenem (IMP 10µg), erythromycin (E 15µg (Bioanalyse LTD, Turkey)), were placed on MHA plates and the diameters of inhibition zones formed follow-ing 24-hour incubation at 37°C were measured [18]. Inter-pretation of the response of Bacillus to these antibiotics was presented as resistant, intermediate or sensitive, based on the size of the inhibition zones for individual antibiotics according to the instructions given in the manufacturer’s manual.
2.4 Determination of antimicrobial activity
The antimicrobial activity of Bacillus strains isolated from soil was tested against 27 microbial genera including 14 bacteria (Bacillus cereus ATCC 7064, Staphylococcus.
epidermidis ATCC 12228, S. aureus ATCC 25923, B. sub-tilis NRRL B-200, MRSA (clinic isolate), Enterococcus fecalis ATCC 29112, Enterobacter aerogenes ATCC
13048, Pseudomonas aeroginosa ATCC 27853, Proteus
vulgaris NRRL B-123, Escherichia coli NRRL 3704, Aer-omonas hydrophilia NRRL 406, VRE (clinic isolate), Aci-netobacter spp. (clinic isolate), Moraxella catarrhalis
(clinic isolate), 3 yeasts (Saccharomyces cerevisa (wild),
S. boulardii (wild), and Rhodotorula rubra DSM 70403),
and 2 mold species (Aspergillus flavus NRRL 1957, and A.
fumigatus NRRL 163). The Bacillus strains were grown in
300mL Erlenmeyer flasks containing 50mL SG (pepton 20.0g, glycerol 20.0mL, tap water 1000mL) medium, with a rotary shaker at 160rpm at 37°C. The SG medium con-sisted of 2% glycerol and 2% peptone in tap-water, pH 7.5 [19]. After allowing maximum biomass formation for
3 days in SG medium, the cultures were centrifuged. The obtained supernatant was passed through 0.22µm Milli-pore filter. All supernatants were maintained at +4°C until they could be used for the agar well diffusion method. The determination of the inhibitory effect of the extracts from isolates on test microorganisms was carried out according to the agar well diffusion method. All bacteria were cul-tured on NB medium and incubated at 37°C for 24 hours. MHA medium (15mL) was poured into each sterile Petri-dish. A suspension (100μL) containing 108CFU/mL for bacteria, 107CFU/mL for yeasts, or 105 spores/mL for molds was spread on the plates of MHA or Sabouraud dex-trose agar medium respectively. Suspensions (100μL) of the target strain cultured for 24 hours were spread on the plates, and wells of 10mm diameter were punched in the agar with a sterile steel borer. The extracts (100μL) were poured into the wells and the plates were incubated at 37°C for 24 hours for bacterial strains, 48 hours for yeasts and at room temperature for 72 hours for fungi.
2.5 Screening for amylase-producing strains
Bacterial isolates were screened for amylolytic proper-ties using a starch hydrolysis test on starch agar plate. The microbial isolates were streaked as a line on the starch agar plate and plates were incubated at 37oC for 24 hours. The plates were flooded with a 1% prepared iodine solution at the end of incubation. A clear zone of hydrolysis surround-ing the growth indicates a positive result while the presence of a blue color around the growth indicates a negative result [20].
2.6 Screening of protease-producing strains
Bacterial isolates were screened for extracellular pro-tease production by streaking onto skim milk agar plates. The plates were incubated at 37oC for 24 hours. Protease production was demonstrated by the clearing of opaque milk proteins in the area surrounding the colony [20].
2.7 PHB producing Bacillus Detection
Microscopic techniques were used to detect PHB-pro-ducing strains. The strain Sudan Black was used to identify PHB under bright field microscopes. Sudan black was placed on heat-fixed samples and prepared as 0.3g Sudan Black B dissolved in 70mL 95% ethanol, bringing it to 100mL with distilled water. Samples were stained with Su-dan Black solution for 10 minutes, dried with filter paper and clarified using xylene drops, dried again with filter pa-per and counterstained with 0.5% aqueous safranine for 5 seconds [21].
3. RESULTS AND DISCUSSION
A total of 27 isolates were obtained from the Kuriki Mound, approximately 1km south-west of the village of Oymataş at the confluence of the Tigris River (Fig 1). Soil samples were taken from different locations; these being the silo, the furnace ruins and graves within the excavation.
The Bacillus was isolated from aquatic environments [22], various parts of insects [23] and grain textures [24].
The results of the Gram stain, endospore stain and cat-alase tests, as well as the morphological properties of the isolates, revealed that all of them belonged to the Bacillus genus. Identification of the 27 isolates was achieved ac-cording to biochemical tests and the VITEK identification system.
According to the biochemical properties, 27 isolates were identified as these being Brevibacillus borstelensis (8 isolates), B. cereus (7 isolates), B. subtilis (6 isolates), B.
macerans (1 isolate), B. popillia (1 isolate), B. polymyxa (1
isolate), B. coagulans (2 isolates), B. larvae (1 isolate). Ten
Bacillus cereus strains were isolated from different soil
samples. Bacillus cereus is found frequently as a sapro-phyte in soil, water, vegetation, and air. The colonization of different ecological niches is enabled by its extremely good adaptability and resistance to various influences [25]. The microbial level of resistance or tolerance of each concentration of heavy metal was depicted by the level of growth on the agar. The microbial load decreased with an increase in the concentration. In the present study, re-sistance to nine heavy metals Ni2+, Zn2+, Co2+, Fe 3+, Cr6+, Pb2+, Mn2+, Cu2+ and Cd +2 was investigated in all the iso-lates. The MICs of the isolates ranged from 0.0625mM/mL to 16mM/mL. Among all the isolates, resistance to heavy metals was determined in comparison to the reference strain (B. subtilis NRRL B-209), (Table 1). Which achieved the following results: Mn2+ [A2 (B. cereus), A4, A7 (B. borstelensis), C1 (B. subtilis), C4 (B. cereus), D4 (B. polymyxa, E3 (B. larvae)] 11.17% and Cd +2 [A2, A3, A5 (B. cereus), A11 (B. borstelensis] 8.51%. The Bacillus isolates were not found to be resistant to other heavy met-als. Çolak et al. [26] reported that isolated Paenibacillus
polymyxa was found to be tolerant to different
concentra-tions of heavy metals, as evidenced by its MICs ranging from 25µg/mL to 1600µg/mL. In assessing the range of MICs obtained, a maximum MIC (1600µg/mL) was ob-served for Mn and Cu, with a minimum for Cd (25µg/mL). However, the control strain showed a maximum MIC of 1600µg/mL for Mn and a minimum of 25µg/mL for Cd and Cu. A heavy metal resistance pattern of Cu=Mn > Pb>Ni=Zn>Hg was observed in P. polymyxa. A metal-re-sistant microbial community was likely to be dependent on the presence of the metal in the growth medium [27]. In this study, the soil sample used for the isolation of bacteria contained serpentine soil. Çolak et al. [28] reported that they isolated B. cereus and B. pumilus from a serpentine soil and found that both isolates were resistant to copper and lead metals. Retaining suitable concentrations of es-sential metals such as copper, while rejecting toxic metals like lead and cadmium was probably one of the toughes challenges of living cells. The first response to toxic metal contamination is a large reduction in microbial activity. This is confirmed by the fact that habitats that have had high levels of metal contamination for years still have mi-crobial populations and activities that are smaller than the
TABLE 1 - Heavy metals resistance profiles of isolated bacteria
Heavy metals n Heavy metal concentrations mM /mL of medium Resistant isolates
0.0625 0.125 0.25 0.5 1 2 4 8 16 n % Ni2+ 27 - - - 3 24 -* - - - 0 0.0 Zn2+ 27 1 3 5 18 - - -* - - 0 0.0 Co2+ 27 1 3 5 18 - -* - - - 0 0.0 Fe 3+ 27 - - - 3 7 17 -* - - 0 0.0 Cr6+ 27 - - - 8 5 14 -* - - 0 0.0 Pb2+ 27 - - 1 7 1 2 16 -* - 0 0.0 Mn2+ 27 - - - - - - 8 12* 7 19 11.17 Cu2+ 27 2 7 18 *- - - - 0 0.0 Cd +2 27 4 *- 23 - - - 23 8.51
* Heavy metals reference strains MIC values
microbial populations in uncontaminated habitats. Konopka et al. (1999) argued that resistance mechanisms do not offer protection at extremely high levels of free metal ions and with a lethal toxic effect [8].
Also there is the use of different types of microorgan-isms such as algae, fungi and bacteria that remove metals from solution. It would necessarily be of immense benefit exploiting microorganisms for this purpose, but the at-tendant health implications that may result when these or-ganisms develop resistant genes invariably becomes a source of concern for disease treatment and management [8].
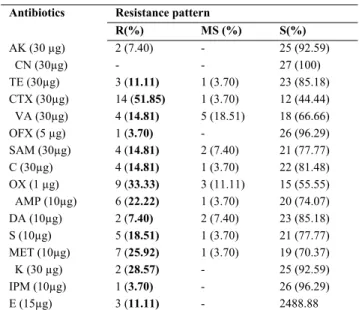
The highest level of antibiotic resistance was observed against cefotaxime while the low level of antibiotic re-sistance was exhibited against gentamycin clindamycin (3.7%), ofloxacine (3.7%), and none of the isolated bacte-ria were resistant to gentamisine (Table 2). Ombui et al. [29] reported that all B. cereus isolates were resistant to ampicillin but susceptible to streptomycin, and that re-sistance to gentamicin was about 7%. Jensen et al. [30] in-vestigated the antimicrobial resistance among B. cereus group isolates from Danish agricultural soil, and recorded the resistance to bacitracin and erythromycin. In this study, all the isolates were resistant to ampicillin and cephotax-ime, but susceptible to kanamycin. However, a differing degree of susceptibility and resistance was observed for other antibiotics tested. It was also reported that since most
Bacillus species populate the same ecosystems as Strepto-myces and other antibiotic producers, they might have
ac-quired resistance to antibiotics produced under natural con-ditions. Since many bacteria exhibit production of second-ary metabolites mainly in the late exponential or in the sta-tionary phase, we investigated whether this is also the case for the production of antimicrobial agent [31]. The antimi-crobial activity of Bacillus strains isolated from the soil was tested against 27 microbial genera, including 14 bac-teria (B. cereus, S. epidermidis, S. aureus, B. subtilis, MRSA (clinic isolate), E. fecalis, E. aerogenes, P.
aer-oginosa, P. vulgaris, E. coli, A. hydrophilia, VRE (clinic
isolate), Acinetobacter spp. (clinic isolate), M. catarhalis (clinic isolate), 3 yeasts (S. cerevisa, S. boulardii, R. rubra),
and 2 mold species (A. flavus, A. fumigatus). The microorgan-isms were tested and the results can be found in Table 3.
TABLE 2 - Antibiotic resistance profiles of isolated bacteria Antibiotics Resistance pattern
R(%) MS (%) S(%) AK (30 µg) 2 (7.40) - 25 (92.59) CN (30µg) - - 27 (100) TE (30µg) 3 (11.11) 1 (3.70) 23 (85.18) CTX (30µg) 14 (51.85) 1 (3.70) 12 (44.44) VA (30µg) 4 (14.81) 5 (18.51) 18 (66.66) OFX (5 µg) 1 (3.70) - 26 (96.29) SAM (30µg) 4 (14.81) 2 (7.40) 21 (77.77) C (30µg) 4 (14.81) 1 (3.70) 22 (81.48) OX (1 µg) 9 (33.33) 3 (11.11) 15 (55.55) AMP (10µg) 6 (22.22) 1 (3.70) 20 (74.07) DA (10µg) 2 (7.40) 2 (7.40) 23 (85.18) S (10µg) 5 (18.51) 1 (3.70) 21 (77.77) MET (10µg) 7 (25.92) 1 (3.70) 19 (70.37) K (30 µg) 2 (28.57) - 25 (92.59) IPM (10µg) 1 (3.70) - 26 (96.29) E (15µg) 3 (11.11) - 2488.88
R: Resistant (≤14), MS: Moderately Sensitive (15-18), S: Sensitive (≥19mm)
Due to the fact that Bacillus species have produced an-tibiotics, in the form of soluble protein, have been found cheaper and more effective in studies conducted so far, therefore these microorganisms are preferred for commer-cial production. It was reported that members ofthe species
Bacillus generally produced polypeptide-type bacteriocins,
and that these antibiotics generally affect Gram-positive bacteria. 27 Bacillus isolates were found most effective against B. cereus S. epidermidis and B. subtilis. Bacillus isolates were found exhibiting antibacterial activity against
B. cereus with zone size ranging from 13-30mm. The Ba-cillus isolates showed no activity against S. aureus strains. Bacillus isolates were also found non-inhibitory against
non-clinical Gram-negative isolates such as E. fecalis, E.
aerogenes, P. aeroginosa, P. vulgaris, E. coli and A. hy-drophila. Oscariz et al. [32] identified and isolated a bac-
TABLE 3 - Identification and some of the characteristic of isolated bacteria
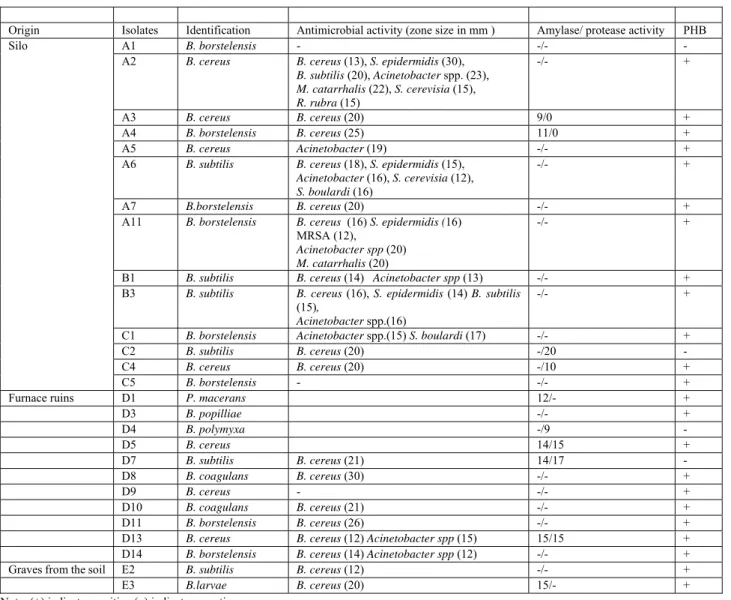
Origin Isolates Identification Antimicrobial activity (zone size in mm ) Amylase/ protease activity PHB
Silo A1 B. borstelensis - -/- -
A2 B. cereus B. cereus (13), S. epidermidis (30),
B. subtilis (20), Acinetobacter spp. (23), M. catarrhalis (22), S. cerevisia (15), R. rubra (15) -/- + A3 B. cereus B. cereus (20) 9/0 + A4 B. borstelensis B. cereus (25) 11/0 + A5 B. cereus Acinetobacter (19) -/- +
A6 B. subtilis B. cereus (18), S. epidermidis (15),
Acinetobacter (16), S. cerevisia (12), S. boulardi (16)
-/- +
A7 B.borstelensis B. cereus (20) -/- +
A11 B. borstelensis B. cereus (16) S. epidermidis (16)
MRSA (12),
Acinetobacter spp (20) M. catarrhalis (20)
-/- +
B1 B. subtilis B. cereus (14) Acinetobacter spp (13) -/- +
B3 B. subtilis B. cereus (16), S. epidermidis (14) B. subtilis
(15),
Acinetobacter spp.(16)
-/- +
C1 B. borstelensis Acinetobacter spp.(15) S. boulardi (17) -/- +
C2 B. subtilis B. cereus (20) -/20 -
C4 B. cereus B. cereus (20) -/10 +
C5 B. borstelensis - -/- +
Furnace ruins D1 P. macerans 12/- +
D3 B. popilliae -/- + D4 B. polymyxa -/9 - D5 B. cereus 14/15 + D7 B. subtilis B. cereus (21) 14/17 - D8 B. coagulans B. cereus (30) -/- + D9 B. cereus - -/- + D10 B. coagulans B. cereus (21) -/- + D11 B. borstelensis B. cereus (26) -/- +
D13 B. cereus B. cereus (12) Acinetobacter spp (15) 15/15 +
D14 B. borstelensis B. cereus (14) Acinetobacter spp (12) -/- +
Graves from the soil E2 B. subtilis B. cereus (12) -/- +
E3 B.larvae B. cereus (20) 15/- +
Note: (+) indicates positive (-) indicates negative
teriocin-producing strain of Bacillus cereus from a soil sample. Wherein the compound, cerein 7, was found active against most Gram-positive bacteria. However, the same compound was reported ineffective Gram-negative bacte-ria. Güven et al. [25]reported that, had moderate activity against diverse Gram-positive bacteria and certain Gramnegative bacteria. Bacillus isolates were found to dis-play antibacterial activity against MRSA, Acinetobacter spp., and M. catarrhalis Bacillus isolates were also found to exhibit antibacterial activity against B. cereus with zones sizes ranging between 13-30 mm, however, they showed no activity against the VRE strain. Three yeasts (S.
cere-visiae, S. boulardii, R. rubra) and 2 fungi (A. fumigatus, A. flavus) showed antifungal activity against the species
eval-uated. Yeast strains showed antifungal activity, but other fungi strains were not inhibitory. The Bacillus species have a wide range of antibacterial activities and are also used as anti-fungals. These results provide us with novel tools for antimicrobial therapy, which is particularly urgent at a time
when many Gram positive pathogens have developed re-sistance mechanisms to almost all known antibiotics. In ad-dition, there is a need to develop an antifungal agent with fewer side effects, since serious fungal infections, such as invasive aspergillosis, have increased dramatically in re-cent years. The A2, A3, D5, D13 (B. cereus), A4 (B.
bor-stelensis), D1 (P. macerans), D7 (B. subtilis), and E3 (B. larvae) isolates showed amylase activity, with D13 and E3
demonstrating the highest amylase activity with a zone di-ameter of 15mm. The isolates indicating protease activity were found to be A2, C4, D5, D13 (B. cereus), C2, D7 (B.
subtilis) and D4 (B.polymyxa). The Bacillus isolate with
the highest protease activity was determined to be C2with a zone diameter of 20mm. Of the four Bacilli isolated from the fish gut (B1, B2, B3 and B4), only one isolate (Bacillus sp. B1) showed activity for protease, lipase, amylase and cellulase enzymes [20]. The poly-β-hydroxybutyrate (PHB) is a compound accumulated as a cellular energy and carbon reserve by a large variety of bacteria that include
the genera Alcaligenes, Pseudomonas, Rhizobium and
Ba-cillus. It is deposited intracellularly in amorphous state in
inclusions in the cytoplasm and inclusion levels depend on the nutritional (carbon source, C/N ratio, etc.) and the en-vironmental conditions (pH, oxygen, etc.) during growt [33]. Considering the poly-β-hydroxybutyrate PHB pres-ence of isolates, PHB prespres-ence is not found in all of the isolates other than A1 (B. borstelensis), C2, D7 (B. subtilis) and D4 (B. polymyxa) strains (Table 3). Gram positive bac-teria reporthave not been reported to accumulate large amountsof polyhydroxyalkonate and hence have not been considered as potent candidates for industrial production. A number of Bacillus spp. has been reported to accumulate 9-67%dry cell weight PHB.
4. CONCLUSION
Our results show that endospore forming Bacillus mi-croorganisms isolated from archaeological excavations soil (Batman) are a potential new source for antimicrobial sub-stances. The study of different environments throughout the world has yielded a lot of antimicrobial agents that are of great value for the treatment of many infectious diseases. Today, increase in the number of drug-resistant pathogens, particularly the acquired multi-drug resistant strains (bac-teria and yeastest), cause serious public health problem throughout the world [34]. Therefore, the need for antimi-crobial discovery and better treatments of these infections, particularly in hospitals where resistance is immediately life threatening, is growing more urgent [35]. Antimicro-bial substances produced by bacteria seem to play an im-portant role in the bacterial antagonism in soil and aquatic ecosystems [36] and might ensure the predominance of a given strain in a bacterial niche against other bacteria of the same species or against other species [37]. The isolated
Ba-cillus has good antimicrobial activity against the tested
gram positive bacteria, gram negative bacteria and yeast. The heavy metal tolerant soil bacteria are a potential indi-cator of toxicity of heavy metals to other forms of life. In this study it is proved that this high manganese tolerant bacteria confirmed the contamination or earth crust by this metal in the study location of excavations soil. The future prospect lies in the application of this microorganism for purposes like heavy metal redediation and potential use in extracting rare metals from dilute solution or removing toxic metals from industrial effluents [38]. The present studies inform us that the isolated Bacillus spp. has the properties to resist a wide of heavy metal (manganese) and antibiotics (cefotaxime and oxacillin); it may be harmful to human and other living.
The strain of Bacillus was found to be a potential pro-ducer of protease and amylase enzyme activity. Bacillus microorganisms have become an important point of study in the search for potential enzymes, purified and character-ized. This strain might be suitable with important industrial applications and economic advantage. Further experiments to enhanced enzyme production for commercialized pro-
cess is needed. The physiological characteristics of the or-ganism are important for the bioprocess to produce poly-mer. This strain might be employed in the industrial pro-duction of PHA. This study aimed to shed light on the ex-cavations made in Turkey.
ACKNOWLEDGEMENT
This work has been dedicated/ conducted with the head of the Batman Museum Directorate, under the auspices of Ministry of Culture and Tourism, General Directorate for Cultural Heritage and Museums and supported by the Gen-eral Directorate of State Hydraulic Works (DSI).
The authors have declared no conflict of interest.
REFERENCES
[1] Genç, E., Valentini, S. and D’Agostino, A. (2011). Kuriki Mound Excavations of 2009. 32. Excavation Results Meeting, 1. Ankara.142-153.
[2] Genç, E., Valentini, S. and D’Agostino, A. (2012) Kuriki Mound Archaeological Project 2010, A Preliminary Report 33. Excavation Results Meeting, 2. Ankara. 463-479. [3] Genç, E., (2013) Kuriki Mound Excavations of 34. Excavation
Results Meeting, 1. Ankara. 229–240.
[4] Genç, E., Yıldız.Köse, B. and Köse, Ç. (2014) 2012 Kuriki Mound Excavations 35. Excavation Results Meeting, 1. An-kara. 292-302.
[5] Setlow, P. (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. Journal of Ap-plied Microbiology 101:514–525.
[6] Gadd, G.M. (1992). Metals and Microorganisms: A problem of definition. FEMS Microbiol. Lett, 100:197-200. [7] Nies, D.H. and Silver, S., (1995). Ion efflux systems involved
in bacterial metal resistances. J. Ind. Microbial., 14: 186-199. [8] Mgbemena, I,C., Nnokwe, J,C.,Adjeroh, L.A. and
Onyeme-kara, N.N. (2012), Resistance of bacteria isolated from Oto-miri river to heavy metals and some selected antibiotics. Cur-rent Research Journal of Biological Sciences. 4(5): 551-556. [9] Sevgi, E., Coral, G., Gizir, M. and Sangün, M.K. (2010).
In-vestigation of heavy metal resistance in some bacterial strains isolated from industrial soils. Turk. J. Biol. 34, 423-43. [10] Grouda, M.K., Swellam, A.E. and Omar, S.H. (2001)
Produc-tion of PHB by a Bacillus megaterium strain using sugarcane molasses and corn step liquor as sole carbon and nitrogen sources. Microbial Research 156, 201-207.
[11] Valappil, S.P., Misra, S.K., Boccaccini, A.R., Keshavarz, T., Bucke, C. and Roya, I. (2007) Large-scale production and ef-ficient recovery of PHB with desirable material properties, from the newly characterised Bacillus cereus SPV Journal of Biotechnology 132, 251–258
[12] Mannova, R.N. and Sattarova, R.K. (2001). Antibiotics pro-duced by Bacillus bacteria. Chem. Nat. Comp. 37, 117-123. [13] Granum, P.E. (1994). Bacillus cereus and its toxins. J. Appl.
[14] Gulam, R. and Nur Hayati, Y. (1995). Prevalence of Bacillus
cereus in selected foods and detection of enterotoxin using
tecra vıa and bcet rpla. Int. J., Food Microbiol., 25, 131-139. [15] Sneath, P.H.A (1986) Endospore forming gram positive rods
and cocci. Bergeys Manuel Syst. Bacteriol. 2, 1104–1207 [16] Alam M.Z. and Malik A. (2008). Chromate resistance,
transport and bioreduction by Exiguobacterium sp ZM 2 iso-lated from agricultural soil irrigated with tannery effluent, J. Basic Microbiol. 48 (5), 416-420.
[17] Alam M.Z., Ahmad S. and Malik A. (2011) Prevalence of heavy metal resistance in bacteria isolated from tannery efflu-ents and affected soil. Environ. Monit. Assess. 178, 281-291. [18] National Committee for Clinical Laboratory Standards
(NCCLS) (1990) Performance standards for antimicrobial disk susceptibility tests. Approved Standard (M2-A4). National Committee for Clinical Laboratory Standards, Villanova, PA. [19] Sessitsch, A., Kan, F.Y. and Pfeifer, U. (2003). Diversity and community structure of culturable Bacillus spp. populations in the rhizospheres of transgenic potatoes expressing the lytic peptide cecropin B. Appl. Soil. Ecol. 22, 149-158.
[20] Ariole C. N., Nwogu H. A. and Chuku P. W.(2014) Enzymatic Activities of Intestinal Bacteria Isolated from Farmed Clarias
gariepinus International Journal of Aquaculture 4 (18),
108-112.
[21] Lopez-Cortes, A., Lanz-Landozuri, A. and Garcia-Maldonado, J.Q. (2008). Screening and isolation of PHB-producing bacte-ria in a polluted marine microbial mat. Microb. Ecol. 56, 112-120.
[22] Faulkner, D.J. (2000) Fihlights of marine natural products chemistry. Nat. Prod. Rep. 17, 1-6.
[23] Gebhardt, K., Schimana, J., Müîler, J., Fiedier,H.P., Kallen-born, G. H., Holzenkampfer, M., Krastel, P., Zeeck, A., Vater, J., Höltzeî, A., Schmid, G. D., Rheinheimer, J. and Dettner, K. (2002). Screening for biologically active metabo-lites with endosymbiotic bacilli isolated from arthropods. FEMS Microbiology Letters 217 (2), 199–205.
[24] Földes, T., Banhegyi, I., Herpai, Z., Varga, L. and Szigeti, J. (2000). Isolation of Bacillus strains from the rhizosphere of cereals and in vitro screening for antagonisms against phyto-pathogenic, food borne pathogenic and spoilage microorgan-isms. J. Appl. Microbiol., 89, 840-846.
[25] Guven, K., Ilhan, S., Mutlu, M. and Colak, F., (2008). Diver-sity, characterization and antimicrobial activities of Bacillus
cereus strains isolated from soil. Fresen. Environ. Bull., 17
(3), 303-310.
[26] Çolak, F., Olgun, A., Atar, N. and Yazıcıoğlu D. (2013) Heavy metal resistances and biosorptive behaviors of Paenibacillus
polymyxa Batch and column studies. Journal of Industrial and
Engineering Chemistry, 19, 3, 863–869.
[27] Hassen N., Saidi M., Cherif A. Boudabous (1998), Resistance of environmental bacteria to heavy metals. Bioresour. Tech-nol. 64, 7–15.
[28] Çolak F., Atar N., Yazıcıoğlu D., Olgun A. (2011) Biosorption of lead from aqueous solutions by Bacillus strains possessing heavy-metal resistance. Chem. Eng. J. 173, 422-428. [29] Ombui, J.N., Mathenge, J.M., Kimotho, A.M., Macharia, J.K.,
and Nduhiu, G. (1996). Frequency of antimicrobial resistance and plasmid profiles of Bacillus cereus strains isolated from milk. East Afr. Med. J. 73, 380-384.
[30] Jensen, G.B., Hansen, B.M., Eilenberg, J. and Mahillon, J. (2003). Minireview: The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol.,5, 631-640.
[31] Brinkhoff, T., Bach, G., Heidorn, T., Liang, L., Schlingloff, A. and Simon, M. (2004) Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl Environ Mi-crobiol 70, 2560-5.
[32] Oscariz, J.C., Lasa, I. and Pisabarro, A.G. (1999) Detection and characterization of cerein 7, a new bacteriocin produced by Bacillus cereus with a broad spectrum of activity. FEMS Microbiol. Letters 178, 337-341.
[33] Laranja, J.L.Q., Ludevese-Pascual, G.L., Amar, E.C., Sorgeloos, P., Bossier, P. and De Schryver. (2014) Poly-beta-hydroxybutyrate (PHB) accumulatıng Bacillus spp. improve the survival, growth and robustness of penaeus monodon (fab-ricius, 1798) postlarvae, veterinary microbiology. http://dx.doi.org/10.1016/j.vetmic.2014.08.011.
[34] Dopazo, C.P., Lemos, M.L., Lodeiros, C., Bolinches, J., Barja, J.L. and Toranzo, A.E. (1988). Inhibitory activity of antibiotic-producing marine bacteria against fish pathogens. J Appl Mi-crobiol. 56, 97-101.
[35] Michel-Briand, Y. and Baysse, C. (2002). The pyocins of
Pseudomonas aeruginosa. Biochimie 84, 499-510.
[36] Darabpour, E., Ardakani, M.R., Motamedi, H., Ghezelbash, G., and Ronagh, M.T. (2010), Isolation of an antibiotic pro-ducer Pseudomonas sp. from the Persian Gulf. Asian Pacific J Trop Med. 3, 318-21.
[37] Shlaes, D.M., Projan, S.J. and Edwards, J.E. (2004). Antibiotic discovery: state of the state. ASM News. 70, 275-81. [38] Samanta, A., Bera, P., Khatu, M., Sinha, C., Pal, P., Lalee, A.
and Mandal, A. (2012). An investigation on heavy metal tol-erance and antibiotic resistance properties of bacterial strain acillus sp. Isolated from municipal waste, Journal of Microbi-ology and BiotechnMicrobi-ology Research 2(1), 178-189.
Received: January 12, 2015 Revised: March 20, 2015 Accepted: April 15, 2015 CORRESPONDING AUTHOR Dr. Ferdağ ÇOLAK Department of Biology Faculty of Science and Art Dumlupınar University Kütahya TURKEY Phone: +90 274 2652031/3152 Fax: +90 274 2652014 Email: fercolak@gmail.com.tr