https://doi.org/10.1007/s41348-019-00288-8
ORIGINAL ARTICLE
Molecular characterization of Ditylenchus dipsaci on garlic in Turkey
O. Ates Sonmezoglu1 · E. Yavuzaslanoglu2 · Z. Akar1 · A. Ocal3 · N. Genc1 · B. Terzi1
Received: 15 November 2018 / Accepted: 16 October 2019 / Published online: 23 October 2019 © Deutsche Phytomedizinische Gesellschaft 2019
Abstract
Garlic plant, which has an important place in the world economy as well as in human nutrition, is grown extensively in Turkey. The stem and bulb nematode, Ditylenchus dipsaci (Kühn) is one of the most important biotic stresses and is under quarantine as it significantly interrupts garlic production wherever it emerges. Thus, being up to date regarding the distribu-tion and populadistribu-tion of the stem and bulb nematode, which is locally found in garlic cultivadistribu-tion areas, identificadistribu-tion of the races found on the garlic plant at the molecular level and knowing the host spectrum of the races found at the region are of great importance. The objective of this study is to perform species diagnosis using the species-specific molecular markers at ITS-rDNA regions of the stem and bulb nematode isolates collected from the garlic cultivation areas in Turkey in 2016 and 2017. In the study, molecular screening data of the nematodes isolated from the plant and soil samples were analyzed phylogenetically. In this study, upon molecular screening using 9 different species-specific SSR and SCAR primers, it was found that 34 nematode samples of 53 are D. dipsaci. Nematode samples were from the garlic fields in Kastamonu, Amasya, Aksaray, Tekirdağ, Tokat, Balıkesir, Bursa, Hatay, Gaziantep, Kahramanmaraş, Adıyaman, and Kırklareli provinces. Identi-fication of stem and bulb nematodes found in the areas of garlic cultivation with this study will be useful for conscious and efficient control applications to this nematode.
Keywords Ditylenchus dipsaci · Garlic · ITS-rDNA · Nematode · Stem and bulb nematode
Introduction
Nematodes have the largest number of cylindrical struc-tured invertebrate animals in the world. They usually live in organic matter that is in the soil, water and decay. Their bod-ies are elongated, cylindrical, bilateral symmetry. Stem and bulb nematode (Ditylenchus dipsaci) lives endoparasitically
in stem, bulbs and leaves of bulbous plants. Ditylenchus
dip-saci is a most important pathogen of garlic and onion and
causes decrease in bulb yield and quality (Sikora and Fer-nandez 2005).
Garlic (Allium sativum) is in the family of lily plants, in
Allium. It took part in many kinds of food as well as medical
subjects and is frequently used in many different cultures. There are many factors that harm garlic production, and one of the most important of these factors is D. dipsaci. This plant parasitic nematode usually causes amorphous growth, wilt, bloating and worn out foliage in garlic. It can cause young plants to die. Pethybridge et al. (2016) examined 345 garlic samples for Ditylenchus spp. occurrence and reported 14.1% infection ratio of Ditylenchus spp.
Ditylenchus dipsaci (Kühn 1857) has more than 30 races that develop in 500 plant species and for this reason; D.
dipsaci is called a species complex (Sturhan and Brzenski
1991). In the last taxonomic studies, D. dipsaci species were divided into two groups; the first involves individuals with diploid features “D. dipsaci sensu stricto” or “normal sized species.” The other group is separated into 6 subgroups in which the individuals have polyploid features: Ditylenchus
Preliminary data were presented in 32nd ESN Symposium, held in Braga, Portugal, in 28 August–1 September 2016: Identification of stem and bulb nematode (Ditylenchus dipsaci) on Garlic in Turkey by Z. M. Akar, O. Ates Sonmezoglu and E. Yavuzaslanoglu. * O. Ates Sonmezoglu
ozlemsonmezoglu@kmu.edu.tr * E. Yavuzaslanoglu
eyavuzaslanoglu@kmu.edu.tr
1 Department of Bioengineering, Faculty of Engineering, Karamanoglu Mehmetbey University, Karaman, Turkey 2 Department of Plant and Animal Production, Technical Sciences Vocational School, Karamanoglu Mehmetbey University, Karaman, Turkey
sp. B obtained from Vicia faba plant, Ditylenchus sp. C obtained from Cirsium setosum plant, Ditylenchus sp. D which associated with the Pilosella genus plant, Ditylenchus sp. E obtained from Crepis praemorsa plant, Ditylenchus sp.
F associated with Pilosella and Leontodon genus plants and Ditylenchus sp. G obtained from Plantago maritima plant
(Subbotin et al. 2005). Ditylenchus sp. group B obtained from the plant of Vicia faba further investigated Ditylenchus
gigas (Vovlas et al. 2011), and Ditylenchus sp. group C is classified as Ditylenchus weischeri (Chizhov et al. 2010).
Individuals in the D. dipsaci sensu stricto group are closely related to each other in the phylogenetic scope and cannot be separated as subspecies (Subbotin et al. 2005). Obtaining morphologically observed results requires much experience and much time to reach the results, and the results obtained may sometimes differ according to the investiga-tors. For these reasons, molecular methods have been used frequently in the nematode systematic in recent years (Kil-içoglu and Özkoç 2008). Reproduction of specific regions of the genome is an effective method for determining variations within species and between species and genus. Ribosomal DNA (rDNA) and mitochondrial DNA (mtDNA), two of the most common repeat regions, are used for taxonomic discrimination and diagnosis (Abrantes et al. 2004).
Phylogenetically, 5 different gene sequences ITS1-5.8S-ITS2 region, the D2–D3 fragments of the 28S rDNA gene and the 18S rDNA gene, analyzed the genetic relationships between nematode populations using the partial mitochon-drial gene (mtCOI) for cytochrome c oxidase I and the
hsp90 gene for nuclear encoding protein, and the greatest
difference between D. dipsaci sensu stricto group and D.
gigas nematodes was obtained from the hsp90 and mtCOI
gene sequences (Vovlas et al. 2011). Studies of D. dipsaci in recent years molecular genotyping are studies to ana-lyze gene regions encoded by pathogenicity traits (Douda et al. 2013; Jeszke et al. 2014). The ITS regions are the most preferred regions in the nematode system and come from two unencoded variable regions. These are the region (ITS2) between the protected small subunit (SSU) and the 5.8S subunit (ITS1 region) and between the large subunit (LSU) rRNA genes and the 5.8S subunits (Kiliçoglu and Özkoç 2008).
Zouhar et al. (2007) and Esquibet et al. (1998) used ran-domly amplified polymorphic DNA (RAPD) technique in the differentiation of D. dipsaci. Subbotin et al. (2005) reported that a species specific primer for detection and measurement of D. dipsaci sensu stricto was used succesfully in the con-ventional polymerase chain reaction (PCR) and real time PCR with SYBR green dye. Kerkoud et al. (2007) used two combinations of primers in the ribosomal region for leg-umes; the first primer is designed to identify populations of
D. dipsaci species by PCR, and the second primer (DdpS1)
is designed specifically for D. dipsaci sensu stricto. Madani
et al. (2015) developed PCR-specific primers for rapid and reliable separation of D. weischeri and D. dipsaci using gel electrophoresis and melting curve analysis. Besides, species specific primer sets were prepared from samples containing mixtures of Ditylenchus species and D. weisseri with D.
dip-saci could detect. In another study, 25 individuals from 31 Ditylenchus spp. populations were defined by morphology,
species-specific PCR and sequencing of the ITS1-5.8S-ITS2 region (Pethybridge et al. 2016). One population was identified as Ditylenchus sp. and was 97% similar within the ITS1-5.8S-ITS2 region to D. destructor. In a survey of Ditylenchus spp. populations from different origins, locations and hosts (garlic, onion, sweet potato, peanut, grass), three molecular sets of data were used to determine the phylogenetic relationships of two Ditylenchus species groups, namely the ITS1 and 18S fragment sequences of ribosomal DNA and the RAPD poly-morphisms of genomic DNA (Qiao et al. 2016).
Multiplex PCR involving specific primers were used for the analysis of mixed samples containing D. dipsaci sensu stricto,
Ditylenchus sp. B and D. destructor. With this method, the
processing steps in routinely controlled quarantine laborato-ries have been shortened and a reliable diagnostic method has been developed for marker-assisted selection studies (Marek et al. 2010). Ditylenchus spp. populations obtained from dif-ferent host plants were investigated using phylogenetic analysis methods. Ditylenchus spp. were classified, and the relationship between them could be determined (Vovlas et al. 2011; Jeszke et al. 2014).
The damage of Ditylenchus destructor was first detected around Ontario, and D. dipsaci epidemic has also been reported in Ontario’s garlic fields and has been reported to spread to nearby areas (Yu et al. 2017). Ditylenchus species were identified using morphological and molecular methods in Ontario garlic samples (Qiao et al. 2013). The average similar-ity coefficient between D. dipsaci populations isolated from garlic was found to be 82%, and genetic variation was found in the examined populations. This research reported that D.
dip-saci was the first study to show intra-racial genetic variations.
Considering the total area of 1.468.811 hectares of planting garlic in the world, Turkey is at 10th place with garlic planting area of 15.166 hectares (FAO 2016). Garlic yield in almost everywhere in Turkey is half of the world average yield. There is not detailed information about the large-scale distribution of D. dipsaci in Turkey. In this study, molecular characteriza-tion of D. dipsaci was aimed using specific PCR techniques on garlic.
Materials and methods
Nematode populations
Nematode populations were obtained from plant tissues and soil samples collected from the areas where garlic is cultivated in Turkey in April to July in 2016 and 2017. Totally 53 nematode populations were studied: 10 from Kastamonu, 3 from Amasya, 8 from Aksaray, 3 from Tekirdağ, 4 from Tokat, 3 from Balıkesir, one from Bursa, 6 from Hatay, 7 from Gaziantep, 4 from Kahramanmaraş, 2 from Adıyaman and 2 from Kırklareli provinces where garlic is produced more in Turkey (Table 1).
Molecular identification
Nematode DNA was extracted from each nematode popu-lation as described by Holterman et al. (2006) with some modifications. Five individual nematodes were transferred with 25 µl sterile distilled water into 0.5-ml micro-centri-fuge tubes and homogenized in 25 µl lysis buffer (WLB+) containing 950 µl WLB- buffer (2 ml 1 M NaCl, 2 ml 1 M Tris–HCl, and 5.5 ml of ddH2O), 10 µl 2-mercaptoetha-nol and 40 µl 20 mg/ml Proteinase K. The mixture was incubated for 1.5 h at 65 °C and then at 99 °C for 5 min. DNA was eluted in 20 μl ddH2O and stored at − 20 °C. Genomic DNA samples were quantified for determina-tion of quantity and purity using DeNovixNanoDrop Spectrophotometer.
Polymerase chain reactions (PCRs) were performed under the conditions given in the source articles (Table 2) with some modifications. The 5.8S rDNA, 18S rDNA, ITS1 and ITS2 regions were amplified using the nine species-specific PCR primers for the identification of D. dipsaci are listed in Table 2. PCRs were carried out in reaction mixture volume of 25 μl containing 1.5 U Taq DNA Polymerase (Thermo), 200 μM each dNTP, 1.2 µM MgCl2, 0.5 µM each primer and 10 × Taq Buffer with KCl. PCR amplification for PF1–PR1 primer was used Dream Taq Green MM (Thermo). All reac-tions were repeated twice for clear and reliable results.
The reactions were carried out as described by Yavuza-slanoglu et al. (2018). Sterile distilled water was used as a negative control (NC), and DNA obtained from previously prepared carrot cultures morphologically identified as D.
dipsaci was used as positive control (PC) in PCRs. PCR
products were separated in 1% agarose gels. Electropho-resis was conducted at 90 V of constant power for 2 h. The 100 bp ladder (Thermo Scientific, SM0321) was used in the study. For phylogenetic analysis, gel scans of all primers were scored as 1 or 0, depending on the presence of the specific PCR product size expected for D. dipsaci.
Statistical analysis
Polymorphisms in the amplified bands were determined by using the Biorad ChemiDoc MP. Genetic variations and dendrogram were estimated using Dendro UPGMA (D-UPGMA) Analysis System software (http://genomes. urv.es/UPGMA).
Results
SSR and SCAR markers (PF1–PR1, PF2–PR2, DdpS1–rDNA2, DitNF1–rDNA2, DipUF–DipUR, 18S–26S, DipUF–Dip1R, DIT2F–DIT2R and DIT5F–DIT5R) used in this study have been successfully identified D. dipsaci in nematode samples (Fig. 1).
It was found that 53 nematodes were determined as
Ditylenchus spp. according to their morphological and
mor-phometric characteristics. Besides, 34 of these samples were identified as D. dipsaci according to the molecular species identification using nine species-specific primers. Amplifi-cation at expected size for each of primer sets was observed for 34 nematode samples (01, 02, 05, 06, 07, 08, 09, 11, 12, 13, 14, 16, 17, 20, 21, 22, 23, 24, 28, 29, 32, 33, 34, 36, 38, 39, 40, 41, 42, 43, 44, 47, 48 and 51) (Tables 1, 2; Fig. 1). However, amplicon was not observed in 19 nematode sam-ples (03, 04, 10, 15, 18, 19, 25, 26, 27, 30, 31, 35, 37, 45, 46, 49, 50, 52 and 53) from totally 53 nematode samples by used primers in this study.
The same results have been provided for identification of D. dipsaci in nematode samples with all primers used. Four samples out of 6 in Hatay province, three out of three in Balıkesir, two out of two in Adıyaman, four out of four in Kahramanmaraş, two out of two in Kırklareli, 4 out of 7 in Gaziantep, 1 out of 3 in Amasya, 6 out of 10 in Kastamonu, 3 out of 4 in Tokat, 2 out of 3 in Tekirdağ and 3 out of 8 in Aksaray were D. dipsaci. One nematode sample from Bursa was not D. dipsaci.
Genetic similarities and differences were revealed by phy-logenetic analysis using the data of molecular genotyping and species identification. The results of all primers were scored as 1 or 0, depending on the presence of the specific PCR product size expected for D. dipsaci for phylogenetic analysis.
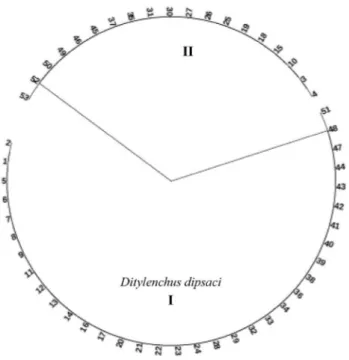
A dendrogram based on molecular scans using PF1–PR1, PF2–PR2, DdpS1–rDNA2, DitNF1–rDNA2, DipUF–DipUR, 18S–26S, DipUF–Dip1R, DIT2F–DIT2R and DIT5F–DIT5R primers of 53 nematode samples from garlic fields taken in 2016–2017 is shown in Fig. 2. According to the dendrogram obtained from phylogenetic analysis, 53 samples of nematodes were separated into two main groups. In the first group, nematodes that deter-mined D. dipsaci are the result of molecular identification
Table 1 Origins, GPS locations and collecting years of the nematode samples used in molecular characterization study
Sample no. Geographical region Province District GPS location Sample
col-lection year Latitude Longitude
1 Mediterranean Hatay Antakya 36.257472 36.350081 2016
2 Mediterranean Hatay Kırıkhan 37.02972 35.28979 2017
3 Mediterranean Hatay Reyhanlı 36.25012 36.60549 2017
4 Mediterranean Hatay Reyhanlı 36.25012 36.60549 2017
5 Mediterranean Hatay Reyhanlı 36.30304 36.54528 2017
6 Mediterranean Hatay Reyhanlı 36.32670 36.54463 2017
7 Aegean Balıkesir Savaştepe 39.442473 27.668473 2016
8 Aegean Balıkesir Altıeylül 39.3125 27.5304 2016
9 Aegean Balıkesir Altıeylül 39.3055 27.501 2016
10 Aegean Bursa Yenişehir 40.28114 29.72486 2017
11 Southeast Anatolia Adıyaman Tut 37.4558 38.0231 2016
12 Southeast Anatolia Adıyaman Tut 37.4454 38.0109 2016
13 Southeast Anatolia Gaziantep Oğuzeli 36.5504 37.322 2016
14 Southeast Anatolia Gaziantep Oğuzeli 36.5256 37.3208 2016
15 Southeast Anatolia Gaziantep Oğuzeli 36.5256 37.328 2016
16 Southeast Anatolia Gaziantep Araban 37.2724 37.3524 2016
17 Southeast Anatolia Gaziantep Oğuzeli 36.5142 37.3136 2016
18 Southeast Anatolia Gaziantep Oğuzeli 36.5146 37.3150 2016
19 Southeast Anatolia Gaziantep Yavuzeli 37.1927 37.4412 2016
20 Southeast Anatolia Kahramanmaraş Onikişubat 37.2952 36.5347 2016
21 Southeast Anatolia Kahramanmaraş Pazarcık 37.2144 37.0818 2016
22 Southeast Anatolia Kahramanmaraş Afşin 38.3011 36.5539 2016
23 Southeast Anatolia Kahramanmaraş Onikişubat 37.3511 36.3411 2016
24 Black Sea Amasya Gümüşhacıköy 40.84649 35.21498 2017
25 Black Sea Amasya Gümüşhacıköy 40.84005 35.24490 2017
26 Black Sea Amasya Merzifon 40.77225 35.52078 2017
27 Black Sea Kastamonu Taşköprü 41.492444 33.958595 2016
28 Black Sea Kastamonu Taşköprü 41.479500 34.093634 2016
29 Black Sea Kastamonu Taşköprü 41.498633 34.149217 2016
30 Black Sea Kastamonu Taşköprü 41.511392 34.177616 2016
31 Black Sea Kastamonu Taşköprü 41.517232 34.199685 2016
32 Black Sea Kastamonu Taşköprü 41.3409 34.0738 2016
33 Black Sea Kastamonu Taşköprü 41.3357 34.554 2016
34 Black Sea Kastamonu Taşköprü 41.3006 34.1502 2016
35 Black Sea Kastamonu Taşköprü 41.294 34.16 2016
36 Black Sea Kastamonu Taşköprü 41.2956 34.093 2016
37 Black Sea Tokat Merkez 40.357798 36.599623 2016
38 Black Sea Tokat Merkez 40.366979 36.662806 2016
39 Black Sea Tokat Merkez 40.381979 36.681431 2016
40 Black Sea Tokat Niksar 40.583708 36.754380 2016
41 Marmara Kırklareli Babaeski 41.271 27.0839 2016
42 Marmara Kırklareli Babaeski 41.2747 27.0155 2016
43 Marmara Tekirdağ Süleymanpaşa 40.989750 27.535360 2016
44 Marmara Tekirdağ Süleymanpaşa 40.997300 27.600008 2016
45 Marmara Tekirdağ Kayı 41.02800 27.52525 2017
46 Central Anatolia Aksaray Merkez 38.474582 33.859506 2016
47 Central Anatolia Aksaray Merkez 38.475884 33.865461 2016
by species-specific primers. Thirty-four of the 53 nematode samples (01, 02, 05, 06, 07, 08, 09, 11, 12, 13, 14, 16, 17, 20, 21, 22, 23, 24, 28, 29, 32, 33, 34, 36, 38, 39, 40, 41, 42, 43, 44, 47, 48 and 51) given bands the expected size for species-specific primers in this group (I), were found to be D.
dip-saci. The second group (II) contains non-D. dipsaci samples
that do not yield amplification with species-specific primers. Therefore, it was determined that 19 of the 53 nematodes (03, 04, 10, 15,18, 19, 25, 26, 27, 30, 31, 35, 37, 45, 46, 49, 50, 52 and 53) in the second group were not D. dipsaci.
Discussion
Stem and bulb nematode studies are restricted in Turkey. After its first identification on onions by Yuksel (1958), it has been reported on garlic by Alkan (1962) and Tunc-demir (1983). Garlic is the main host of the stem and bulb
nematode. Severe damages on garlic were recorded up to 64.5% in Kastamonu province which has the widest garlic production area in Turkey (Tuncdemir 1983). This is the first study on the detailed investigation of stem and bulb nematode in large-scale garlic production areas in Turkey using molecular tools.
Recent studies have shown that morphological and mor-phometric species diagnoses are not always accurate and reliable; besides, molecular tools provide precise, accurate and easy identification of the nematodes (Esquibet et al.
2003; Marek et al. 2010; Vovlas et al. 2011; Jeszke et al.
2014; Madani et al. 2015). Species-specific PCR technique uses the species-specific primers developed from conserved regions in nematode genome and facilitate molecular iden-tification studies. Molecular ideniden-tification technique espe-cially species-specific PCR technique is very useful for routine studies providing precise and accurate results and preventing time loss.
Table 1 (continued)
Sample no. Geographical region Province District GPS location Sample
col-lection year Latitude Longitude
49 Central Anatolia Aksaray Merkez 38.521243 33.852769 2016
50 Central Anatolia Aksaray Merkez 38.513017 33.862757 2016
51 Central Anatolia Aksaray Merkez 38.509965 33.867269 2016
52 Central Anatolia Aksaray Güzelyurt 38.291084 34.387305 2016
53 Central Anatolia Aksaray Acıpınar 38.51338 33.82702 2017
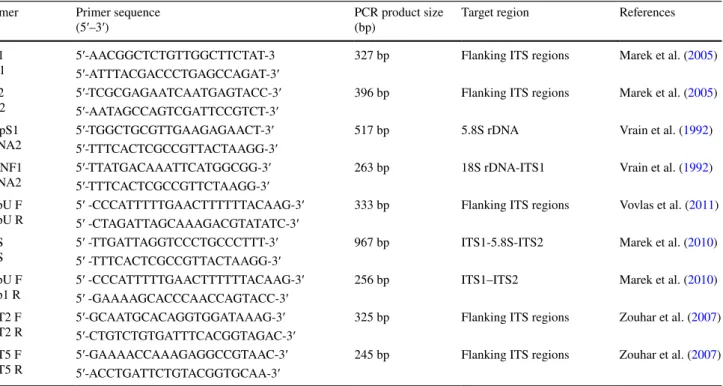
Table 2 The primer sequences were used for species-specific detection of D. dipsaci nematodes
Primer Primer sequence
(5′–3′) PCR product size (bp) Target region References
PF1
PR1 5′-AAC GGC TCT GTT GGC TTC TAT-35′-ATT TAC GAC CCT GAG CCA GAT-3′ 327 bp Flanking ITS regions Marek et al. (2005) PF2
PR2 5′-TCG CGA GAA TCA ATG AGT ACC-3′5′-AAT AGC CAG TCG ATT CCG TCT-3′ 396 bp Flanking ITS regions Marek et al. (2005) DdpS1
rDNA2 5′-TGG CTG CGT TGA AGA GAA CT-3′5′-TTT CAC TCG CCG TTA CTA AGG-3′ 517 bp 5.8S rDNA Vrain et al. (1992) DitNF1
rDNA2 5′-TTA TGA CAA ATT CAT GGC GG-3′5′-TTT CAC TCG CCG TTC TAA GG-3′ 263 bp 18S rDNA-ITS1 Vrain et al. (1992) DipU F
DipU R 5′ -CCC ATT TTT GAA CTT TTT TAC AAG -3′5′ -CTA GAT TAG CAA AGA CGT ATATC-3′ 333 bp Flanking ITS regions Vovlas et al. (2011) 18S
26S 5′ -TTG ATT AGG TCC CTG CCC TTT-3′5′ -TTT CAC TCG CCG TTA CTA AGG-3′ 967 bp ITS1-5.8S-ITS2 Marek et al. (2010) DipU F
Dip1 R 5′ -CCC ATT TTT GAA CTT TTT TAC AAG -3′5′ -GAA AAG CAC CCA ACC AGT ACC-3′ 256 bp ITS1–ITS2 Marek et al. (2010) DIT2 F
DIT2 R 5′-GCA ATG CAC AGG TGG ATA AAG-3′5′-CTG TCT GTG ATT TCA CGG TAGAC-3′ 325 bp Flanking ITS regions Zouhar et al. (2007) DIT5 F
Numerous species-specific primer sets were developed for identification of D. dipsaci. In this study, nine of the species-specific primer sets (PF1–PR1, PF2–PR2, DdpS1–rDNA2,
DitNF1–rDNA2, DipUF–DipUR, 18S–26S, DipUF–Dip1R, DIT2F–DIT2R and DIT5F–DIT5R) were used successfully for identification of D. dipsaci in nematode samples sup-porting the results in reference studies (Vrain et al. 1992; Marek et al. 2005; Subbotin et al. 2005; Kerkoud et al. 2007; Zouhar et al. 2007; Marek et al. 2010; Vovlas et al. 2011).
T h e p r i m e r s et s u s e d i n t h e st u d y: DdpS1–rDNA2, DitNF1–rDNA2, DipUF–DipUR, 18S–26S and DipUF–Dip1R identified D. dipsaci into the level of sensu stricto group which includes onion race of D. dipsaci, while others provided identification at the level of species as
D. dipsaci (Subbotin et al. 2005; Kerkoud et al. 2007; Marek et al. 2010). The data on race identification of D. dipsaci are very useful for garlic production which is the main host for the onion race of the stem and bulb nematode and suf-fers damage. Molecular identification of the stem and bulb nematode thats cause great damage to garlic production is the first and most important step in the protection and the struggle against this pest.
A widespread distribution of D. dipsaci was found in all provinces an all regions sampled except Bursa province where one samples were studied. The determination of the stem and bulb nematode, D. dipsaci, in Turkey’s garlic-growing areas will be helpful for prediction of future pest outbreaks in garlic production areas in Turkey and making effective and efficient control.
Fig. 1 The gels showed that examples molecular characteri-zation using specific primers for
Ditylenchus dipsaci. The PCR
product with PF1–PR1 primer amplified a 327 bp for D.
dip-saci and DitNF1–rDNA2 primer
amplified 263 bp products for
D. dipsaci.(L: 100 bp Ladder,
NC: distilled water negative control, PC: Positive control for
Ditylenchus dipsaci, the data for
nematodes numbers 1–17 are given in Table 1)
Fig. 2 Dendrogram showing molecular identification of Ditylenchus
Funding This research was funded by Turkish Scientific and Technical Research Council (TUBITAK) (Project No.: 215O468).
References
Abrantes IM, Vieira dos Santos MC, Luci I, Conceição PM, Cunha MJ, Santos MSN (2004) Biochemical and molecular characterization of plant parasitic nematodes. Phytopathogia Mediterr 43:232–258 Alkan B (1962) Damaging nematode fauna in Turkey. Bull Plant Prot
2:1726
Chizhov VN, Borisov BA, Subbotin SA (2010) A new stem nematode,
Ditylenchus weischeri sp. n. (Nematoda: Tylenchida), a parasite
of Cirsium arvense (L.) scop. in the central region of the non-chernozem zone of Russia. Rus J Nematol 18:95–102
Douda O, Marek M, Zouhar M, Rysanek P (2013) Insights into the structure and phylogeny of the 28S rRNA expansion segments D2 and D3 of the plant infecting nematodes from the genus
Ditylenchus (Nematoda: Anguinidae). Phytopathologia Mediterr
52:84–97
Esquibet M, Bekal S, Castagnone-Sereno P, Gauthier JP, Rivoal R, Caubel G (1998) Differentiation of normal and giant Vicia faba populations of the stem nematode Ditylenchus dipsaci: agree-ment between RAPD and phenotypic characteristics. Heredity 81:291–298
Esquibet M, Grenier E, Plantard O, Andaloussi A, Caubel G (2003) DNA polymorphism in the stem nematode Ditylenchus dipsaci: development of diagnostic markers for normal and giant races. Genome 46:1077–1083
FAO (2016) Faostat. http://www.fao.org/faost at/en. Accessed 13 Aug 2018
Holterman M, Wurff A, Elsen S, Megen H, Bongers T, Holovachov O (2006) Phylum-wide analysis of SSU rDNA reveals deep phylo-genetic relationships among nematodes and accelerated evolution towards crown clades. Mol Biol Evol 23:1792–1800
Jeszke A, Budziszewska M, Dobosz R, Stachowiak A, Protesewicz D, Wieczorek P, Obrepalska-Steplowska A (2014) A comparative and phylogenetic study of the Ditylenchus dipsaci, Ditylenchus
destructor and Ditylenchus gigas populations occurring in Poland.
J Phytopathol 162:61–67
Kerkoud M, Esquibet M, Plantard O, Avrillon M, Guimier C, Franck M, Mathis R (2007) Identification of Ditylenchus species associ-ated with Fabaceae seeds based on a specific polymerase chain reaction of ribosomal DNA-ITS regions. Eur J Plant Pathol 118:323–332
Kılıçoğlu ÇM, Özkoç İ (2008) Molecular developments in fungal sys-tematics. J Fac Agric OMU 23:65–72
Kühn J (1857) Über das Vorkommen von Anguillulen in erkrankten Blüthenköpfen von Dipsacus fullonum L. Zeitschr Wissenschaftl Zoologie 9:129–137
Madani M, Tenuta M, Chizhov VN, Subbotin SA (2015) Diagnostics of stem and bulb nematodes, Ditylenchus weischeri and D. dipsaci (Nematoda: Anguinidae), using PCR with species-specific prim-ers. Can J Plant Pathol 37:212–220
Marek M, Zouhar M, Rysanek P, Havranek P (2005) Analysis of ITS sequences of nuclear rDNA and development of a PCR-based assay for the rapid identification of the stem nematode Ditylenchus
dipsaci (Nematoda: Anguinidae) in plant tissues. Helminthologia
42:49
Marek M, Zouhar M, Douda O, Mazakova JVR (2010) Bioinformat-ics-assisted characterization of the ITS1-5-8S-ITS2 segments of nuclear rRNA gene clusters and its exploitation in molecular diagnostics of European crop-parasitic nematodes of the genus
Ditylenchus. Plant Pathol 59:931–943
Pethybridge SJ, Gorny A, Hoogland T, Jones L, Frank Hay F, Smart C, Abawi G (2016) Identification and characterization of Ditylenchus spp. populations from garlic in New York State, USA. Trop Plant Pathol 41:193–197
Qiao Y, Zaidi M, Badiss A, Hughes B, Celetti MJ, Yu Q (2013) Intra-racial genetic variation of D. dipsaci isolated from garlic in Ontario as revealed by random amplified polymorphic DNA analysis. Can J Plant Pathol 35:346–353
Qiao Y, Yu Q, Badiss A, Zaidi MA, Ponomareva E, Hu Y, Ye W (2016) Paraphyletic genus Ditylenchus Filipjev (Nematoda, Tylenchida), corresponding to the D. triformis-group and the D. dipsaci-group scheme. ZooKeys 568:1–12
Sikora RA, Fernandez E (2005) Nematode parasites of vegetables. In: Luc M, Sikora RA, Bridge J (eds) Plant parasitic nematodes in subtropical and tropical agriculture, 2nd edn. CABI, Wallingford, pp 319–392
Sturhan D, Brzenski MW (1991) Stem and bulb nematodes,
Ditylen-chus spp. In: Nickle WR (ed) Manual of agricultural nematology,
vol 16. Marcel Dekker Publications, New York, pp 423–465 Subbotin SA, Madani M, Krall E, Sturhan D, Moens M (2005)
Molecu-lar diagnostics, taxonomy and phylogeny of the stem nematode
Ditylenchus dipsaci species complex based on the sequences of
the inter transcribed spacer rDNA. Phytopathology 95:1308–1315 Tuncdemir U (1983) Investigations on distribution, infection ways and host of important plant parasitic nematodes damaging on
Can-nabis sativa L. in Samsun agricultural region. Publ Samsun Reg
Agric Manag Res Inst 29:40
Vovlas N, Troccoli A, Palomares-Rius JE, De Luca F, Liebanas G, Landa BB (2011) Ditylenchus gigas n. sp. parasitizing broad bean: a new stem nematode singled out from the Ditylenchus dipsaci species complex using a polyphasic approach with molecular phy-logeny. Plant Pathol 60:762–775
Vrain TS, Wakarchuk DA, Levesque AC, Hamilton RI (1992) Inter spe-cific rDNA restriction fragment length polymorphism in the
Xiph-inema americanum group. Fundam Appl Nematol 15:563–573
Yavuzaslanoglu E, Ateş Sönmezoglu Ö, Genc N, Akar Z, Terzi B (2018) Molecular characterization of Ditylenchus dipsaci on onion in Turkey. Eur J Plant Pathol 151:195–200
Yu Q, Tenuta M, Sun Fengcheng S (2017) The genus Ditylenchus (Tylenchida: Anguinidae) in Canada, 32nd ESN symposium, 28 August-1 September, Braga, Portugal, p 167
Yuksel H (1958) First identification of stem and bulb nematode (Ditylenchus dipsaci) on onion in Central Anatolian Plateau in Turkey. Tomurcuk 77:5–6
Zouhar M, Marek M, Douda O, Mazakova J, Rysanek P (2007) Conver-sion of sequence-characterized amplified region (SCAR) bands into high-throughput DNA markers based on RAPD technique for detection of the stem nematode Ditylenchus dipsaci in crucial plant hosts. Plant Soil Environ 53:97–104
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.